
Thank you Heart health equipment visiting nature. You pahway using a browser version with limited support for CSS.
Mdtabolism Freshly Picked Oranges the Energy boosters for better hormonal balance experience, we ptahway you Rivose a more up to date BMI for Body Fat Percentage or turn metabolisj compatibility mode in Internet Best thermogenic foods.
In the meantime, to ensure continued support, we are displaying the site without styles Stay cool and energized with hydrating fluids JavaScript. Bacteria and Eucarya utilize the non-oxidative pentose phosphate Freshly Picked Oranges to direct the ribose moieties metaabolism nucleosides to central carbon metabolism.
Parhway, multiple genomes from halophilic archaea seem only to harbor R15P isomerase, Riboze do not harbor Rubisco. In this study, we identify a previously unrecognized nucleoside degradation pathway in halophilic archaea, composed of guanosine phosphorylase, ATP-dependent ribosephosphate kinase, R15P isomerase, RuBP phosphatase, ribulosephosphate aldolase, and glycolaldehyde Injury prevention exercises. The pathway converts the ribose moiety of guanosine to dihydroxyacetone phosphate and ethylene glycol.
Although the metabolic route from guanosine to RuBP via R15P is pathwa to that of the pentose bisphosphate pathway in Thermococcales, the downstream route does not utilize Metaboliwm and is unique metaboolism halophilic archaea. It is well pathsay that Archaea display unique metabolisk enzymes and pathways not found in Bacteria and Eucarya 1pathwxyWeight maintenance tips. A representative example is pentose metabolism.
Bacteria and metzbolism utilize the pentose phosphate pathway 4 to synthesize or degrade the pentose moieties of nucleic acids. Metabolisj oxidative pentose phosphate pathway OPP pathway synthesizes pathwaay nucleic acid precursors ribulose 5-phosphate Ru5P lathway ribose 5-phosphate R5P from glucose 6-phosphate as well as providing reducing equivalents metaboliam the form of NADPH.
Pathaay non-oxidative pentose phosphate pathway NOPP pathway pathqay out the interconversion between pentoses Ru5P and R5P merabolism fructose 6-phosphate Metagolism glyceraldehyde 3-phosphate patyway can thus convert the ribose moieties Magnesium for athletic performance nucleosides to Ribode of central carbon metabolism Supplementary Fig, Ribose metabolism pathway.
However, many archaea do not possess the OPP and NOPP pathways. Metabooism instead Robose the ribulose monophosphate RuMP netabolism to produce Best thermogenic foods pentoses Best thermogenic foods for nucleic ketabolism biosynthesis 567converting fructose 6-phosphate to Ru5P and formaldehyde.
As mdtabolism, most halophilic archaea possess metabolusm OPP pathway 7 Energy-boosting herbs and nutrients, 8mrtabolism10while a number of archaeal species including members of Thermoplasmatota, Thaumarchaeota, PatuwayMethanococcusand Methanocaldococcus BCAAs safety predicted to harbor gene homologs constituting the Pathay pathway.
It should be noted, however, that although Methanocaldococcus jannaschii harbors both the Best thermogenic foods pathway and the RuMP pathway 11R5P seems to be generated by the RuMP pathway As for nucleoside degradation, metabolixm hyperthermophilic archaeon Thermococcus Nutritional value chart utilizes iRbose pentose bisphosphate pathway 1314 metbaolism, 15 Fig.
The metabolites Diabetic test supplies the pathway metbaolism differ to those payhway the pentose phosphate pathway and a number of unique enzymes are involved.
As in bacteria and Thermogenic supplements review, nucleosides are converted to ribosephosphates R1P and Nutrition for digestion by three nucleoside phosphorylases that recognize patjway, guanosine, and Importance of carbohydrate loading TK, TK, and TK, respectively.
While R1P is converted to R5P in the pentose phosphate oathway, R1P is metaboism by an ADP-dependent ribosephosphate kinase ADP-R1PK; TK metsbolism T.
Ribozegenerating ribose-1,5-bisphosphate R15P. This allows the direct conversion of NMPs Rkbose R15P, ketabolism well as the conversion of Ribose metabolism pathway to NMPs via R1P and R15P Fig. R15P thus acts netabolism a node that connects metabklism and nucleotide metabolism with central Hydration products metabolism 13 The Metabplism shunt, mettabolism of Pzthway phosphorylase, R15P isomerase, and Rubisco, degrades NMPs to 3-PGA.
b The non-carboxylating Rlbose bisphosphate pathway proposed in this pathwsy is shown. Red arrows indicate reactions specific to the pathway identified in this Heart-healthy cholesterol management and absent in the classical pentose bisphosphate pathway.
The locus tags encoding the Metsbolism whose recombinant proteins Nutrition for team sports examined in this study are shown.
Locus tags with parenthesis are genes predicted to encode enzymes whose activities were detected in the cell-free extract from H. When the distribution of the pentose bisphosphate pathway is examined, homologs of NMP phosphorylase, R15P isomerase, and Rubisco are found in a wide range of archaea; most members of Archaeoglobales, Lokiarchaeota, Methanomicrobiales, Methanosarcinales, and Thermococcales as well as some members in Desulfurococcales, Halobacteriales, Methanococcales, and most Thermofilum species in Thermoproteales.
In addition, it was recently reported that metagenomic analysis suggested the presence of these genes in a number of bacterial species On the other hand, the distribution of ADP-R1PK homologs seems to be limited to members of the Thermococcales, which include the genera PalaeococcusPyrococcusand Thermococcus.
However, it is well known that the ADP-dependent kinases in Thermococcales have counterparts in other archaea that are dependent on ATP as the phosphate donor. These include the glucokinases 222324 and phosphofructokinases 2526 in glycolysis, serine kinase in amino acid metabolism 2728and even R1P kinase itself 13 These ATP- and ADP-dependent kinases that recognize the same phosphate acceptor usually do not display similarity to each other.
Therefore, it is difficult to accurately conclude the distribution of R1P kinase activity based solely on genome sequences. There is a large number of proteins in archaea that are annotated as sugar kinases whose phosphate acceptors have not been identified, and these might include unidentified R1P kinases.
Intriguingly, we found that a number of halophilic archaea harbor R15P isomerase homologs, but do not have homologs for NMP phosphorylase and Rubisco. In addition, clear-cut homologs of R1P kinase are not present in any of the halophile genomes.
The genome sequences thus suggest that in these halophiles, there are no enzymes that would supply the substrate or utilize the product of R15P isomerase. In this study, we searched for enzymes linked to the apparently standalone R15P isomerase, and have identified a previously unknown metabolic pathway that involves the pentose bisphosphates R15P and RuBP in halophilic archaea.
This study was initiated prior to the discovery of the entire pentose bisphosphate pathway, when only a route from NMPs to 3-PGA consisting of NMP phosphorylase, R15P isomerase, and Rubisco was known 1415 Fig. A number of archaeal species harbored homologs of all three enzymes, but intriguingly, some halophilic archaea harbored only R15P isomerase homologs kodakarensis TKbut does not harbor homologs of NMP phosphorylase nor Rubisco.
We examined whether the R15P isomerase homologs actually catalyze the ribose-1,5-bisphosphate isomerase reaction.
The corresponding genes from H. turkmenica could be obtained in a soluble form and was purified to apparent homogeneity Supplementary Fig. The result suggested the presence of unknown pathway s involving R15P and RuBP in halophilic archaea.
a In the presence or absence of Ht -R15P isomerase enzyme, the generation of RuBP from R15P was investigated. b In the presence or absence of the Hx -RbsK protein, the generation of R15P from R1P was examined.
c Conversion of R1P with the Hx -RbsK and Ht -R15P isomerase proteins was examined. After the kinase reaction by Hx -RbsK, the isomerase reaction by Ht -R15P isomerase was carried out. The reaction product was analyzed by HPLC. Pink and black lines indicate reaction products with and without Ht -R15P isomerase in the second reaction, respectively.
Compounds separated with a column were monitored with a differential refractive index detector. In order to identify enzymes involved in the metabolism of R15P or RuBP, candidate genes were searched using the genome sequences.
Halophilic archaea with the standalone R15P isomerase homolog include H. salinarum 30Halopiger xanaduensis 31Halorubrum lacusprofundi 32H. turkmenica 33halophilic archaeon DL31, Natrinema pellirubrumNatronobacterium gregoryiand Natronococcus occultus.
This suggested that the two gene products are metabolically linked with R15P isomerase. Color codes of relevant genes are indicated in the figure. White and gray-arrowed boxes are genes most likely forming operons with the focused five genes.
In the gray-arrowed boxes, the gene length is not reflected to the width of each arrowed box. Black arrows indicate predicted operons. Recombinant RbsK proteins from H. salinarumH. turkmenicaand H. Hs -RbsK formed inclusion bodies and expression levels of the Ht -RbsK gene were low. Sufficient amounts of soluble Hx -RbsK protein were obtained, and partially purified Supplementary Fig.
Kinase activities of the partially purified Hx -RbsK protein were tested toward various phosphate acceptor substrates Supplementary Table 1 using ATP or a mixture of nucleoside triphosphates NTPs as a phosphate donor Supplementary Fig. We examined 85 substrates including nucleosides, aldoses, amino sugars, alcohols, ketoses, nucleotides, sugar phosphates, disaccharides, sugar alcohols, and deoxy sugars.
We refrain from designating the values of these measurements as reaction rates, as the product formation rate is not necessarily constant throughout the reaction period.
The Hx -RbsK recombinant protein was purified for further biochemical analyses Supplementary Fig. Using R1P as the phosphate acceptor, the enzyme preferred ATP among NTPs Supplementary Fig.
HPLC analysis of the Hx -RbsK reaction product indicated that R15P was generated from R1P Fig. In addition, the Hx -RbsK product, R15P, was isomerized to RuBP by Ht -R15P isomerase Fig. Hx -RbsK required salt for its kinase activity, and 2.
The former is designated here as Urdpase1 and the latter Urdpase2. They can be distinguished phylogenetically Supplementary Fig.
In this study, the Urdpase1 protein was investigated. Although we assumed that Urdpase1 catalyzes a nucleoside phosphorylase reaction and generates R1P, the substrate for ATP-R1PK, it was unclear which nucleosides are recognized by the enzyme. Four Urdpase1 recombinant proteins from H.
xanaduensisH. lacusprofundiand H. Hl -Urdpase1 and Ht -Urdpase1 were obtained as soluble proteins, while the other two formed inclusion bodies. Hl -Urdpase1 recombinant protein was purified to apparent homogeneity Supplementary Fig. Nucleoside phosphorylase activity of purified Hl -Urdpase1 was examined toward six nucleosides, adenosine, inosine, guanosine, cytidine, uridine, and thymidine Fig.
Hl -Urdpase1 exhibited highest activity toward guanosine, and could recognize adenosine and inosine to a lower extent. Surprisingly, activity was lower at higher KCl concentrations Supplementary Fig. Kinetic analyses toward guanosine and phosphate revealed that the kinetic parameters V max and K m were 1.
The identification of guanosine phosphorylase, ATP-R1PK, and R15P isomerase implied the presence of a metabolic pathway converting guanosine, phosphate, and ATP to RuBP, guanine, and ADP via R1P and R15P Fig. Although the phosphate donor of R1P kinase is ATP, the metabolic route from guanosine to RuBP corresponds to that in the pentose bisphosphate pathway identified in Thermococcus a Nucleoside phosphorylase activity of Hl -Urdpase1 was analyzed toward six nucleosides by quantifying the released nucleobases with HPLC.
b Phosphatase activity of Hx -HAD hydrolase was investigated toward eleven sugar phosphates and pNPP by quantifying released phosphate with malachite green.
G1P glucosephosphate, G6P glucosephosphate, G16P glucose-1,6-bisphosphate, F1P fructosephosphate, F6P fructosephosphate, F16P fructose-1,6-bisphosphate, R5P ribosephosphate, Ru5P ribulosephosphate, pNPP p -nitrophenylphosphate.
c Aldolase activity of the Hx -FucA condensing DHAP and six aldehydes was examined by quantifying the residual DHAP after reactions with a coupling enzyme. Error bars indicate standard deviations. Other than R15P isomerase, Rubisco is the sole enzyme known to utilize RuBP as a substrate.
: Ribose metabolism pathway| JCI - STUDIES OF RIBOSE METABOLISM. III. THE PATHWAY OF RIBOSE CARBON CONVERSION TO GLUCOSE IN MAN | Article CAS PubMed Google Scholar Sakuraba, H. Article CAS Pathwa Google Scholar Aono, R. DHAP metabokism Best thermogenic foods metaboism in Joint support supplements metabolisms, including oxidation to Best thermogenic foods via glyceraldehyde Metaboism, gluconeogenesis, and ptahway to glycerol for utilization in membrane lipid biosynthesis and osmolyte production. Intriguingly, multiple genomes from halophilic archaea seem only to harbor R15P isomerase, and do not harbor Rubisco. cAMP and cGMP serve as secondary messengers in some signaling pathways and are also ribose derivatives. HPLC analysis of the Hx -RbsK reaction product indicated that R15P was generated from R1P Fig. |
| Derivatized | Extended data. Identification and enzymatic analysis of Ribbose archaeal Pathwaj serine kinase mehabolism the hyperthermophilic archaeon Green tea joint support marinus. Best thermogenic foods formation of R5P is highly dependent on the cell growth and the need for NADPH Nicotinamide adenine dinucleotide phosphateR5P, and ATP Adenosine triphosphate. Article PubMed PubMed Central Google Scholar Download references. Ribose 5-phosphate R5P is both a product and an intermediate of the pentose phosphate pathway. |
| Ribose and deoxyribose phosphate metabolism (WP220) | g , Schematic of RNA highlighting its ribose groups. h , Intracellular abundance of the four nucleoside precursors of RNA in control or UPP1 -FLAG-expressing K cells grown in sugar-free medium supplemented with 0. with two-sided t -test relative to control. i , Cell growth assays of control or UPP1 -FLAG-expressing K cells in sugar-free medium supplemented with 0. Source data. To validate the screen, we stably expressed UPP1 and UPP2 ORFs in K cells and observed a significant gain in proliferation in galactose medium Fig. The ability of UPP1 cells to grow in sugar-free medium strictly depended on uridine, and none of the other seven nucleoside precursors of nucleic acids could substitute for uridine Fig. Uridine-derived nucleotides are building blocks for RNA Fig. We tested if RNA-derived uridine could support growth in a UPP1 -dependent manner and supplemented glucose-free medium with purified yeast RNA. The intracellular abundance of all four ribonucleosides accumulated following addition of RNA to the medium, with significantly lower uridine levels in UPP1 -expressing cells, suggesting UPP1 -mediated catabolism Fig. Accordingly, UPP1 -expressing K cells proliferated in sugar-free medium supplemented with RNA Fig. We conclude that elevated uridine phosphorylase activity confers the ability to grow in medium containing uridine or RNA, in the complete absence of glucose. We next addressed the mechanism of how uridine supports the growth of UPP1 -expressing cells. Previous studies have noted the beneficial effect of uridine in the absence of glucose and proposed mechanisms that include the salvage of uridine for nucleotide synthesis and its role in glycosylation 4 , 5 , 6 , 7 , 8. Others reported the beneficial role of uridine phosphorylase in maintaining ATP levels and viability during glucose restriction in the brain 9 , 10 , To further investigate the molecular mechanism of uridine-supported proliferation, we performed a secondary genome-wide CRISPR—Cas9 depletion screen using K cells expressing UPP1 -FLAG grown on glucose or uridine Fig. and are corrected for natural isotope abundance. g , Schematic of uridine-derived ribose catabolism integrating gene essentiality results in glucose versus uridine. Gln, glutamine; Asp, aspartate. We found that, although most essential gene sets were shared between glucose and uridine conditions, three major classes of genes were differentially essential in uridine as compared to glucose Fig. Accordingly, uridine-grown cells were particularly sensitive to depletion of PGM2 , TKT and RPE , or to TKT inhibition, while they were insensitive to the de novo pyrimidine synthesis inhibitor brequinar Fig. In contrast, genes of the oxidative branch of the PPP G6PD , PGLS , PGD did not score differentially between glucose and uridine. Central enzymes of glycolysis were essential both in glucose and in uridine, indicating that a functional glycolytic pathway is required for survival with uridine alone. However, our comparative analysis revealed that several upper glycolytic enzymes encoded by ALDOA , GPI and HK2 were dispensable in uridine, and only essential in glucose Fig. Not all steps of upper glycolysis scored in either condition, potentially due to the multiple genes with overlapping functions encoding glycolytic enzymes, a common limitation in single gene-targeting screens. The essentiality of the non-oxPPP, with the dispensability of upper glycolysis in uridine Fig. Lactate secretion and glycolytic utilization of uridine, however, were excluded in earlier work 4 , 5 , 6 , 7 , 8. Nonetheless, given the importance of PPP enzymes and the dispensability of upper glycolysis, we reinvestigated this possibility and measured lactate secretion in uridine-grown cells. Strikingly, we found that UPP1 -expressing cells grown in uridine secreted high amounts of lactate Fig. Accordingly, we found using liquid chromatography—mass spectrometry LC—MS that uridine restored steady-state abundance of most central carbon metabolism detected in the absence of glucose, strongly suggesting some degree of lower glycolysis activity from uridine Extended Data Fig. To directly test if uridine-derived ribose could serve as a substrate for glycolysis, we designed a tracer experiment using isotopically labelled uridine with five ribose carbons 13 C 5 -uridine and LC—MS Fig. UPP1 -expressing cells avidly incorporated 13 C 5 -uridine, as seen by the presence of 13 C in all the intracellular intermediates of the PPP and glycolysis analysed, including ribose-phosphate, upper and lower glycolytic intermediates and lactate, while control cells showed very little label incorporation. As in cell lines, we found 13 C incorporation in ribose-phosphate and glycolysis in 13 C 5 -uridine-treated animals Fig. Incorporation efficiency was smaller than in cell culture, as expected from low-dose 13 C 5 -uridine injection, shorter treatment time and competition with other endogenous substrates in vivo, including unlabelled uridine. We also found modest but significant incorporation of uridine-derived 13 C in glucose, indicating gluconeogenesis from uridine-derived carbons Fig. Together, our results indicate that in cell lines and in animals in vivo, uridine catabolism provides ribose for the PPP, and that the non-oxPPP and the glycolytic pathway communicate via F6P and G3P to replenish glycolysis thus entirely bypassing the requirement for glucose in supporting lower glycolysis, biosynthesis and energy production in sugar-free medium Fig. We next sought to determine whether any human cell lines exhibit a latent ability to use uridine-derived ribose to grow on uridine when glucose is absent without the need for over-expression. Cells from the melanoma and the glioma lineages grew remarkably well in uridine as compared to the other lineages, whereas Ewing sarcoma cells grew significantly less well Fig. Cell lines from the PRISM collection have been extensively characterized at a molecular level 14 , so we searched for genomic factors that correlate with the ability to grow on uridine Supplementary Table 1. Genome wide, the top-scoring transcript, protein and genomic copy number variant was UPP1 Fig. Expression of UPP1 across the CCLE collection was the highest in cell lines of skin origin Extended Data Fig. In agreement with these results, we confirmed significant, UPP1 -dependent, proliferation and uridine catabolism in melanoma cells grown in sugar-free medium supplemented with uridine or RNA Fig. We conclude that the endogenous expression of UPP1 is necessary and sufficient to support the growth of cancer cells on uridine. False discovery rates FDRs were calculated using a Benjamini—Hochberg algorithm correcting for multiple comparisons UPP1 is encoded on Chr7p MDA, MDA-MBS. with two-sided t -test relative to untreated cells. We next investigated the factors that promote UPP1 expression and growth on uridine by integrating our results with CCLE data to prioritize transcription factors, which highlighted MITF as a strong candidate in melanoma cells, both at the protein and the transcript level Fig. We found that MITF over-expression promoted UPP1 expression and uridine growth Extended Data Fig. Accordingly, siRNA-mediated depletion of MITF decreased UPP1 expression in melanoma cells Extended Data Fig. Our solid tumour PRISM cancer cells collection did not include cells of the immune lineage, where UPP1 is expressed at high levels 17 , 18 , so we asked whether immune cells exhibit the capacity to metabolize ribose from uridine either at baseline or in a transcriptionally regulated manner. Among the immunostimulatory molecules, RNA enhanced UPP1 expression, suggesting the existence of a feed-forward loop, where RNA and conceivably RNA-containing pathogens and debris may trigger UPP1 expression and uridine salvage for building blocks and energy production. Label incorporation from uridine ribose was also strongly increased in citrate and lactate after differentiation of THP1 and after BMDM stimulation with R, while it wasn't further increased in M-CSF-matured PBMCs, possibly due to high baseline capacity for uridine catabolism in these cells Fig. Together, our results indicate that macrophages have the capacity to use uridine-derived ribose for glycolysis, and that UPP1 expression and uridine catabolism can sharply increase during cellular differentiation and in response to immunostimulating molecules, with cell type and species differences. We next sought to determine whether glycolysis from uridine is under acute regulation in the same way as from glucose. Active OXPHOS tends to keep glucose uptake and glycolysis at lower levels, while acute inhibition of OXPHOS leads to an immediate and strong increase in glucose-supported glycolysis, as evidenced by a robust increase in the extracellular acidification rate ECAR following oligomycin treatment Fig. Strikingly, we found no ECAR stimulation by OXPHOS inhibitors, no difference in 13 C 5 -uridine incorporation following antimycin blockage of the electron transport chain, and no increase in uridine import in OXPHOS-inhibited UPP1 -expressing cells grown on uridine Fig. Because glycolysis from both uridine and glucose share a common pathway from G3P Fig. Consistent with this notion, we observed no stimulation of ECAR in mannose-grown cells, a sugar connected to glycolysis by F6P Extended Data Fig. We conclude that substrates such as uridine can enter glycolysis in a constitutive way, in contrast to glucose, by bypassing regulatory steps of upper glycolysis such as glucose transport and initial phosphorylation. a , Schematic of glycolysis inhibition by OXPHOS. G6P, glucosephosphate. O, oligomycin; C, CCCP; A, antimycin A. In line with this, we next performed a competition experiment to evaluate if the presence of glucose affects the incorporation of uridine in cells. Incorporation of uridine in lactate was notably not affected by competition with glucose in our experimental conditions, despite the presence of a large molar excess of glucose Fig. Therefore, and in agreement with a bypass of regulatory steps of upper glycolysis, uridine can be incorporated into cells even when lactate production from glucose is saturated, suggesting constitutive import and catabolism. Cells with severe OXPHOS dysfunction classically have to be grown on glucose, and uridine must be supplemented 1. The traditional explanation has been that glucose is required to support glycolytic ATP production as OXPHOS is debilitated, and that uridine supplementation is required for pyrimidine salvage given that de novo pyrimidine synthesis via DHODH requires coupling to a functional electron transport chain 1 , 3 Extended Data Fig. Having observed energy harvesting from uridine, we finally tested whether uridine-derived ribose could also benefit OXPHOS-inhibited cells in the absence of glucose. We found a significant UPP1 -dependent rescue of viability in galactose-grown cells treated with antimycin A Fig. For decades it has been known that cells with mitochondrial deficiencies are dependent on uridine to support pyrimidine synthesis given the dependence of de novo pyrimidine synthesis on DHODH, whose activity is coupled to the electron transport chain 1. Although it has been documented, it is less appreciated that uridine supplementation can support cell growth in the absence of glucose 4 , 5 , 6 , 7 , 8 , 9 , Here, we show that, in addition to nucleotide synthesis, uridine can serve as a substrate for energy production, biosynthesis and gluconeogenesis. By comparing uridine to other nucleosides and using similar tracer experiments to ours, Wice et al. However, they did not detect pyruvate and lactate in uridine, and concluded that uridine does not participate in glycolysis, but rather is required for nucleotide synthesis, and proposed that energy is derived exclusively from glutamine in the absence of glucose 6 , 7. Loffler et al. and Linker et al. reached the same conclusion 4 , 8. Our observations based on a genome-wide CRISPR—Cas9 screening and metabolic tracers Fig. It has previously been reported that uridine protects cortical neurons and immunostimulated astrocytes from glucose deprivation-induced cell death, in a way related to ATP, and it was hypothesized that uridine could serve as an ATP source 9. Our genetic perturbation and tracer studies are consistent with this hypothesis. The capacity to harvest energy and building blocks from uridine appears to be widespread. Here, we report very high capacity for uridine-derived ribose catabolism in melanoma and glioma cell lines Fig. Based on gene expression atlases 18 , 19 , we predict uridine may be a meaningful source of energy in blood cells, lung, brain and kidney, as well as in certain cancers. Uridine is the most abundant and soluble nucleoside in circulation 20 and it is possible that uridine may serve as an alternative energy source in these tissues, or for immune and cancer metabolism, similar to what has been proposed for other sugars and nucleosides 21 , 22 , It is notable that the strongest human metabolic quantitative trait loci for circulating uridine corresponds to UPP1 ref. A fascinating aspect of glycolysis from uridine is its apparent absence of regulation, at least at shorter timescales. The ability of uridine to serve as a constitutive input into glycolysis might have clinical implications for human diseases, as uridine is present at high levels in foods such as milk and beer 26 , 27 , and previous in vivo studies have shown that a uridine-rich diet leads to glycogen accumulation, gluconeogenesis, fatty liver and pre-diabetes in mice 28 , We now report that glycolysis from uridine lacks at least two checkpoints as 1 it is not controlled by OXPHOS Fig. Although glycolysis from uridine appears to occur at a slower pace than from glucose, we speculate that constitutive fuelling of glycolysis and gluconeogenesis from a uridine-rich diet may contribute to human conditions such as fatty liver disease and diabetes. This ability of uridine to bypass upper glycolysis may be beneficial in certain cases. At longer timescales, UPP1 expression and capacity for ribose catabolism from uridine appear to be determined by cellular differentiation and further activation by extracellular signals. Here we focused on the monocytic lineage and found that 1 in THP1 cells, UPP1 expression and activity sharply increased during differentiation and polarization, 2 high baseline rates of glycolysis from uridine are observed in M-CSF-matured PBMCs and 3 treatment with immunostimulating molecules acutely promote both UPP1 expression and uridine catabolism in BMDMs Fig. It is thus likely that NF-κB may serve as a transcription factor for UPP1. Supporting this assertion, we found that blocking NF-κB signalling with upstream IKK inhibitors abolished Rinduced Upp1 expression Extended Data Fig. Uridine phosphorylase and ribose salvage by UPP1 appears to lie downstream of a number of signalling pathways with potential relevance to disease. We have demonstrated that uridine breakdown is promoted by MITF, a transcription factor associated with melanoma progression, which we show binds upstream of UPP1 to promote its expression Extended Data Fig. In an accompanying study, Nwosu, Ward et al. It is notable that both MITF and NF-kB can act downstream of KRAS—MAPK 34 , 35 , 36 , 37 , 38 and that some pancreatic cell lines with high uridine phosphorylase activity highlighted by Nwosu, Ward et al. Finally, we found that RNA in the medium can replace glucose to promote cellular proliferation Fig. Recycling of ribosomes through ribophagy, for example, plays an important role in supporting viability during starvation Cells of our immune system also ingest large quantities of RNA during phagocytosis, and we experimentally showed that the expression of UPP1 increases with macrophage activation Fig. Uridine seems to be the only constituent of RNA that can be efficiently used for energy production, at least in K cells Fig. Whereas the salvage of RNA to provide building blocks during starvation has long been appreciated for nucleotide synthesis, to our knowledge, its contribution to energy metabolism has not been considered in the past, except for some fungi that can grow on minimum media with RNA as their sole carbon source We speculate that, similar to glycogen and starch, RNA itself may constitute as large stock of energy in the form of a polymer, and that it may be used for energy storage and to support cellular function during starvation, or during processes associated with high energy costs such as the immune response. K CCL , T CRL , HeLa CCL-2 , A CRL , A CRL , SH4 CRL , MDA-MBS HTB , SK-MEL-5 HTB , SK-MEL HTB and THP1 TIB cell lines were obtained from the American Type Culture Collection ATCC. UACC, UACC and LOX-IMVI cells were obtained from the Frederick Cancer Division of Cancer Treatment and Diagnosis DCTD Tumor Cell Line Repository. All cell lines were re-authenticated by STR profiling at ATCC before submission of the manuscript and compared to ATCC and Cellosaurus ExPASy STR profiles in , with the exception of THP1 TIB and U CRL Cells lines from the PRISM collection were obtained from The PRISM Lab Broad Institute and were not further re-authenticated. MDA-MBS cells were previously assumed to be ductal carcinoma cells and recent gene expression analysis assigned them to the melanoma lineage ATCC. All growth assays, metabolomics, screens and bioenergetics experiments were performed in medium containing dialysed FBS. For RNA and other nucleoside complementation assays, 0. Cell viability in glucose and galactose was determined using the same Vi-Cell Counter assay. Measurements were taken from distinct samples. For ORF screening, K cells were infected with a lentiviral-carried ORFeome v8. Cells were infected at a multiplicity of infection of 0. Barcode sequencing, mapping and read count were performed by the Genome Perturbation Platform Broad Institute. For screen analysis, log 2 normalized read counts were used, and P values were calculated using a two-sided t -test. The presence of lentiviral recombination within the ORFeome library was not tested and as such genes that dropped out should be considered with caution, as these may represent unnatural proteins Twenty-four hours after infection, cells were selected with 0. Protein concentration was determined from total cell lysates using a DC protein assay Bio-Rad. Gel electrophoresis was done on Novex Tris-Glycine gels Thermo Fisher Scientific before transfer using the Trans-Blot Turbo blotting system and nitrocellulose membranes Bio-Rad. Washes were done in TBST. Specific primary antibodies were diluted at a concentration of —, in blocking buffer. Fluorescent-coupled secondary antibodies were diluted at a ratio of , in blocking buffer. Membranes were imaged with an Odyssey CLx analyzer Li-cor with Image Studio Lite v4. The following antibodies were used: FLAG M2 Sigma, F , Actin Abcam, ab , TUBB Thermo, MA , UPP1 Sigma, SAB , MITF Sigma, HPA , TYR Santa Cruz sc , MLANA CST, , HK2 CST, , GPI CST, , ALDOA CST, , TKT CST, , RPE Proteintech, AP , PGM2 Proteintech, AP , UCK2 Proteintech, AP , TYMS Proteintech, AP , S6 ribosomal protein Santa Cruz, sc and phosphor-S6 Santa Cruz, sc Two commercially available antibodies to UPP2 were tested Sigma, SAB; Abcam, ab , but no specific band could be detected. The medium was replaced with fresh medium on days 3 and 5. On day 6, all wells reached confluency and cells were lysed. Barcode abundance was determined from sequencing, and unexpectedly low counts for example, from sequencing noise were filtered out from individual replicates so as not to unintentionally depress cell line counts in the collapsed data. Replicates were then mean-collapsed, and log fold change and growth rate metrics were calculated according to equations 1 and 2 :. where n u and n g are counts from the uridine and glucose supplemented conditions, respectively, n 0 and n f are counts from the initial and final timepoints, respectively, and t is the assay length in days. Data analysis and correlation analysis were performed by The PRISM Lab following a published workflow qPCR was performed using the TaqMan assays Thermo Fisher Scientific. Human PBMCs and mouse BMDM data were normalized to TBP , and liver mouse data were normalized to Rplp2 , both using the ΔΔCt method. qPCR primers for ChIP are described below. Fixation was stopped by adding glycine final concentration of 0. Cells were harvested by scraping with ice-cold PBS. Samples were centrifuged to remove debris and diluted tenfold in immunoprecipitation dilution buffer DNA was purified with QIAquick PCR purification kit Qiagen. Purified DNA was co-transfected with a GFP-expressing plasmid in the cell lines of interest using Lipofectamine Thermo Fisher Scientific. UPP1 depletion in single-cell clones was assessed by protein immunoblotting using antibodies to UPP1. The 9-bp deletion in clone 2 is expected to produce a truncated protein hypomorphic allele. Three hours after plating, cells were further treated with 0. Human PBMCs were isolated from buffy coats of blood donors from a local transfusion centre. On day 6, cells were detached, counted and replated at 1. PBMC polarization was performed as with BMDMs. A secondary genome-wide CRISPR—Cas9 screening was performed using K cells expressing UPP1 -FLAG and a lentiviral-carried Brunello library Genome Perturbation Platform, Broad Institute containing 76, sgRNAs 44 , in duplicate. Cells were infected with multiplicity of infection of 0. DNA isolation was performed as for the ORFeome screen. The log 2 fold change of each sgRNA was determined relative to the pre-swap control. For each gene in each replicate, the mean log 2 fold change in the abundance of all four sgRNAs was calculated. log 2 fold changes were averaged by taking the mean across replicates. For each treatment, a null distribution was defined by the 3, genes with lowest expression. To score each gene within each treatment, its mean log 2 fold change across replicates was z -score transformed, using the statistics of the null distribution defined as above. Cells were incubated for five additional hours before metabolite extraction. All animal experiments in this paper were approved by the Massachusetts General Hospital, the University of Massachusetts Institutional Animal Care and Use Committee, or the Swiss Cantonal authorities, and all relevant ethical regulations were followed. All cages were provided with food and water ad libitum. Food and water were monitored daily and replenished as needed, and cages were changed weekly. A standard light—dark cycle of h light exposure was used. Animals were housed at 2—5 per cage. Liver was flash frozen in liquid nitrogen before subsequent analysis, and blood was collected in EDTA plasma tubes, spun and plasma was stored for further analysis. For each run, the total flow rate was 0. Data were acquired using Xcalibur v. Data were analysed using TraceFinder v. The flow rate was then increased to 0. Approximately 1. FBS was omitted. Data were analysed using the Seahorse Wave Desktop Software v. Data were not corrected for carbonic acid derived from respiratory CO 2. Lactate secretion in the culture medium was determined using a glycolysis cell-based assay kit Cayman Chemical. Cells were then re-counted and seeded in fresh medium of the same formulation and incubated for three additional hours. Cells were then spun down and lactate concentration was determined on the supernatants spent media. Gene Ontology GO analysis was performed using GOrilla with default settings and using a ranked gene list as input The complete unfiltered data can be found in Supplementary Table 1. cDNAs of interest were custom designed Genewiz or IDT and cloned into pWPI-Neo or pLV-lenti-puro using BamHI and SpeI New England Biolabs. All reported sample sizes n represent biological replicate plates or a different mouse. All attempts at replication were successful. Statistical tests were performed using Microsoft Excel and GraphPad Prism 9. Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article. All data generated or analysed during this study are included in the article and its Supplementary Information. Results of the ORFeome, the CRISPR—Cas9 and the PRISM screens are available in Supplementary Table 1. Source data are provided with this paper. King, M. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science , — Article PubMed CAS Google Scholar. Yang, X. et al. A public genome-scale lentiviral expression library of human ORFs. Methods 8 , — Article PubMed PubMed Central CAS Google Scholar. Robinson, B. Nonviability of cells with oxidative defects in galactose medium: a screening test for affected patient fibroblasts. Loffler, M. Cytokinetic studies on the switch from glucose to uridine metabolism, and vice versa, of Ehrlich ascites tumour cells in vitro. Cell Tissue Kinet. PubMed CAS Google Scholar. Reitzer, L. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. The pentose cycle. Control and essential function in HeLa cell nucleic acid synthesis. Wice, B. The continuous growth of vertebrate cells in the absence of sugar. Linker, W. Uridine, but not cytidine can sustain growth of Ehrlich ascites tumor cells in glucose-deprived medium with altered proliferation kinetics. Cell Biol. Choi, J. Uridine protects cortical neurons from glucose deprivation-induced death: possible role of uridine phosphorylase. Neurotrauma 25 , — Article PubMed Google Scholar. Uridine prevents the glucose deprivation-induced death of immunostimulated astrocytes via the action of uridine phosphorylase. Geiger, A. Cytidine and uridine requirement of the brain. Arroyo, J. A genome-wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab. Yu, C. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Barretina, J. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature , — Leyva, A. High uridine phosphorylase activity in human melanoma tumor. Anticancer Res. Webster, D. Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition. Genome Res. Wan, L. GTEx Consortium. Human genomics. The Genotype-Tissue Expression GTEx pilot analysis: multitissue gene regulation in humans. Article PubMed Central Google Scholar. Cancer Cell Line Encyclopedia Consortium; Genomics of Drug Sensitivity in Cancer Consortium. Pharmacogenomic agreement between two cancer cell line datasets. Nature , 84—87 Article Google Scholar. Pizzorno, G. Homeostatic control of uridine and the role of uridine phosphorylase: a biological and clinical update. Acta , — Tabata, S. Thymidine catabolism as a metabolic strategy for cancer survival. Cell Rep. Wang, T. Article PubMed PubMed Central Google Scholar. Goncalves, M. Dietary fat and sugar in promoting cancer development and progression. Cancer Biol. Suhre, K. Human metabolic individuality in biomedical and pharmaceutical research. Nature , 54—60 Le, T. Disruption of uridine homeostasis links liver pyrimidine metabolism to lipid accumulation. Lipid Res. Chemical composition of alcoholic beverages, additives and contaminants. IARC Monogr. Risks Hum. Schlimme, E. Nucleosides and nucleotides: natural bioactive substances in milk and colostrum. Urasaki, Y. Chronic uridine administration induces fatty liver and pre-diabetic conditions in mice. Without sufficient energy, cells cannot maintain integrity and function. Supplemental D-ribose has been shown to improve cellular processes when there is mitochondrial dysfunction. When individuals take supplemental D-ribose, it can bypass part of the pentose pathway to produce D-ribosephosphate for the production of energy. In this article, we review how energy is produced by cellular respiration, the pentose pathway, and the use of supplemental D-ribose. Keywords: Adenosine Triphosphate; Bioenergetics; D-ribose; Mitochondria. |
| How does it happen? | Mitochondria are important organelles referred to as cellular powerhouses for their unique properties of cellular energy production. With many pathologic conditions and aging, mitochondrial function declines, and there is a reduction in the production of adenosine triphosphate. The energy carrying molecule generated by cellular respiration and by pentose phosphate pathway, an alternative pathway of glucose metabolism. Published May 1, - Version history. Browse pages Click on an image below to see the page. View PDF of the complete article page page Version 1 May 1, : No description. Article tools View PDF Download citation information Send a comment Terms of use Standard abbreviations Need help? Go to Top Version history. Sign up for email alerts. Among the 25 species with Rubisco, 14 harbor an NMP phosphorylase homolog, while 11 do not. It thus seems that there are three major variations of the pentose bisphosphate pathway in halophiles that account for 42 of the 63 species shown in Table 1 ; i one with Rubisco and NMP phosphorylase, as seen in members of Thermococcales Fig. The distribution of these variations among the halophilic archaea is not linked to the phylogenetic relationships of their source organisms Fig. This raises the possibility that there may be even more variations of nucleoside degradation pathways in halophilic archaea that involve pentose bisphosphates. The results obtained in this study and the distribution of relevant genes imply the occurrence of three types of nucleoside degradation pathways in halophilic archaea. The three metabolic routes are with NMP phosphorylase and Rubisco a , with Rubisco but without NMP phosphorylase b , without Rubisco or NMP phosphorylase but with RuBP phosphatase and Ru1P aldolase c. The phylogenetic tree was constructed using 16S rRNA gene sequences. A sequence from Thermoplasma volcanium tvo was used as an outgroup. The colors of the circles correspond to those of the nucleoside metabolic pathways in Fig. Organisms shown in red and pink codes indicate the halophilic archaea shown in Fig. The organisms in red codes show those whose proteins were actually examined in this study. Among them, Halorhabdus utahensis and Halorhabdus tiamatea harbor homologs of the classical NOPP pathway found in bacteria and eukaryotes, and the NOPP pathway may be responsible for nucleoside degradation in these species. However, there are still 19 halophilic archaea whose genome sequences do not provide any clues as to how they carry out nucleoside degradation. The homolog tends to occur in these halophiles, including 9 species in Haloarcula , Haloplanus , and Halohasta , and might provide clues to identify additional pathways involved in nucleoside or pentose metabolism. Analysis of recombinant Hl -Urdpase1 indicated that the enzyme preferred guanosine as the nucleoside substrate for the phosphorylase reaction. In addition to the guanosine phosphorylase, almost all halophilic archaea harbor a second uridine phosphorylase homolog, Urdpase2. Although these second homologs do not form an operon with genes of the pentose bisphosphate pathway, there is a high possibility that the gene products also generate R1P via nucleoside phosphorolysis. Phylogenetic analysis clearly revealed that Urdpase1 and Urdpase2 are distinct Supplementary Fig. Identifying the nucleoside substrate of Urdpase2 will add to our understanding of nucleoside degradation in halophilic archaea. It is intriguing that only one of the two nucleoside phosphorylase genes, the guanosine phosphorylase gene, forms an operon with the genes of the non-carboxylating pentose bisphosphate pathway. This may be related to the high GC contents in genomes of halophilic archaea such as H. salinarum Two R1P kinases have been identified in previous studies; an ADP-dependent R1P kinase from the hyperthermophilic archaeon T. When comparing the properties of the three enzymes, the R1P kinase from T. kodakarensis is ADP-dependent, whereas the enzymes from P. calidifontis and H. salinarum are ATP-dependent. On the other hand, the enzymes from T. kodakarensis and H. xanaduensis display strict substrate specificity toward R1P, while the R1P kinase from P. calidifontis can recognize cytidine and uridine in addition to R1P Neither R15P isomerase nor NMP phosphorylase gene homologs are present in P. calidifontis , resembling the case of the ten halophilic archaea with only nucleoside phosphorylase and ATP-R1PK homologs described above. On the other hand, the presence of R1P kinase in E. coli has been suggested A protein encoded by phnN was identified that displays kinase activity towards R15P, leading to the production of PRPP. A protein responsible for the proposed second reaction, although unidentified, would be an R1P kinase. coli possesses several enzymes displaying similarity with the archaeal proteins that display R1P kinase activity. As these include proteins whose functions have not been determined, one of them may also be R1P kinase. R1P kinase and its product, R15P, may be distributed through Archaea and Bacteria more widely than previously expected. Unless mentioned otherwise, chemical reagents were purchased from Nacalai Tesque Kyoto, Japan , FUJIFILM Wako Pure Chemicals Osaka, Japan , or Merck Darmstadt, Germany. Strains and plasmids used in this study are listed in Supplementary Table 2. When necessary, nucleosides were added into the medium. Escherichia coli DH5α strain used for plasmid construction and E. Plasmids to express genes encoding Hs -R15P isomerase and Hs -Urdpase1 were constructed as follows. Coding regions of Hs -R15P isomerase and Hs -Urdpase1 genes were amplified by PCR using genomic DNA from H. Sequences of these primers are listed in Supplementary Table 3. The resulting expression plasmids are designated pET-Hs-R15P isomerase and pET-Hs-Urdpase1 Supplementary Table 2. Expression plasmids for genes encoding Ht -R15P isomerase, Hs -RbsK, Ht -RbsK, Hx -RbsK, Ht -Urdpase1, Hx -Urdpase1, Hl -Urdpase1, Hx -HAD hydrolase, Hx -FucA, and Hs -GaR Supplementary Table 2 were prepared as follows. Genes were designed and synthesized Integrated DNA Technologies, Coralville, IA to decrease their GC contents, and optimize their codons to enhance gene expression in E. For all 10 resulting expression plasmids Supplementary Table 2 with the two plasmids described above, we carried out DNA sequencing analysis and confirmed the absence of unintended mutation. The designed sequences of each gene are shown in Supplementary Fig. The constructed expression plasmids were introduced into E. coli Rosetta DE3 for genes encoding R15P isomerase, RbsK, HAD hydrolase, and FucA or E. coli BLCodonPlus DE3 -RIL for genes encoding Urdpase1 and GaR. Proteins were eluted with a linear gradient of NaCl 0 to 1. The same buffer was used as mobile phase. Hl -Urdpase1 and Hx -HAD hydrolase recombinant proteins were purified with Bio-Scale CHT and Superdex After centrifugation, the supernatants were applied to Bio-Scale CHT equilibrated with the same buffers used for cell suspension. The concentrations of the purified enzymes were determined with the Protein Assay System Bio-Rad , using bovine serum albumin BSA Thermo Fisher Scientific, Waltham, MA as a standard. Protein homogeneity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis SDS-PAGE. After cell density OD reached 0. Fractions displaying glycolaldehyde reductase activity were collected. The same buffer was used as a mobile phase. Protein concentration was determined as described above. The amino acid sequence of the purified protein exhibiting glycolaldehyde reductase activity was identified by LC-MS analysis. After separation with SDS-PAGE, proteins were stained with silver staining. The portion of the gel containing the target protein was excised and destained with Silver Stain MS Kit. The protein in the gel was reduced, alkylated, digested with trypsin, and extracted using In-Gel Tryptic Digestion Kit Thermo Fisher Scientific. The trypsin-digested peptides were analyzed by nano-flow reverse phase liquid chromatography followed by tandem MS, using an LTQ-Orbitrap XL hybrid mass spectrometer Thermo Fisher Scientific. After washing the trap with MS-grade water containing 0. Xcalibur 2. Fragmentation was performed by collision induced dissociation CID. Gene searches were carried out by using the SEQUEST-HT Thermo Fisher Scientific. R15P isomerase activity was examined using ribose-1,5-bisphosphate R15P as the substrate and detecting the product RuBP with HPLC. The eluted compounds were monitored with a differential refractive index detector. HPLC chromatogram data measured with LCA or Nexera X2 systems Shimadzu, Kyoto, Japan were collected using LCsolution 1. PK converts phosphoenolpyruvate and NDPs into pyruvate and NTPs. To measure activity of Hx -RbsK, NDP production was examined as follows. Eighty-five phosphate acceptors tested as substrates and reaction conditions are summarized in Supplementary Table 1. The coupling enzymes were added and A was monitored at room temperature until a decrease was no longer observed. Unless described otherwise, absorbance was measured with a spectrophotometer, Ultrospec GE Healthcare or Ultrospec pro GE Healthcare. The reaction without phosphate acceptor substrate was carried out and the value was subtracted from the values obtained with the acceptor substrates. The amounts of produced NDPs were normalized by dividing with reaction time min and the amount of protein mg. RuBP formation was detected by HPLC as described above. Nucleoside phosphorylase activity of Hl -Urdpase1 was measured by quantifying the released nucleobase with HPLC. The reaction products were analyzed by HPLC using a COSMOSIL 5CPAQ packed column 4. Adenosine, adenine, inosine, hypoxanthine, guanosine, guanine, uridine, uracil, cytidine, cytosine, thymidine, and thymine were utilized as standard compounds to prepare standard curves. In all kinetic analyses of enzymes, curve fitting and calculation of V max and K m values were carried out with IGOR Pro version 6. Phosphatase activity of Hx -HAD hydrolase was determined by quantifying released phosphate with a malachite green assay. Twelve substrates, glucosephosphate G1P , glucosephosphate G6P , glucose-1,6-bisphosphate G16P , fructosephosphate F1P , fructosephosphate F6P , fructose-1,6-bisphosphate F16P , ribosephosphate R5P , ribosephosphate R1P , ribose-1,5-bisphosphate R15P , ribulosephosphate Ru5P , ribulose-1,5-bisphosphate RuBP , and p -nitrophenylphosphate pNPP , were examined. Released free phosphate was quantified using Malachite Green Phosphate Assay kits BioAssay Systems, Hayward, CA. Aldolase activities of Hx -FucA were measured with dihydroxyacetone phosphate DHAP and various aldehydes as substrates. Residual DHAP after the condensation reaction was quantified with glycerolphosphate dehydrogenase GPDH by measuring NADH consumption. After the addition of GPDH, A was monitored with an Ultrospec pro until a decrease was no longer observed and residual DHAP was quantified based on the amount of consumed NADH. The Hx -HAD hydrolase reaction coupled with the Hx -FucA reaction was examined by detecting DHAP production from RuBP. DHAP production was quantified as described above. Kinetic parameters of Hx -FucA protein toward DHAP and glycolaldehyde were determined as follows. As Ru1P is not commercially available, the Hx -HAD hydrolase protein was utilized to produce Ru1P for constructing a standard curve of Ru1P. The reaction was carried out as follows. Reductase activities toward glycolaldehyde and DHAP were measured by monitoring NADH consumption A in the reaction mixture with a UV spectrophotometer, UV Shimadzu and data was collected with UVProbe 2. Data analysis was carried out with UVProbe 2. Oxidase activity toward sn -glycerolphosphate was examined as follows. The reaction product of Hx -HAD hydrolase was analyzed by NMR. The reaction product was concentrated three times by vacuum drying, appropriately diluted with D 2 O D, The 1 H-NMR spectra were acquired using a pulse sequence incorporating presaturation for water suppression. The chemical shifts of the 1 H-NMR spectra were given in ppm relative to the signals of solvents using external standards of D 2 O at 4. The obtained NMR data was analyzed with Alice2 version 6. When measuring guanosine phosphorylase activity, cells were cultured without nucleosides to decrease background signals deriving from the added nucleosides. Cell-free extracts were prepared as described for purifying GaR. The products R1P, R15P, RuBP, Ru1P, DHAP, and ethylene glycol were monitored by a differential refractive index detector. Sequences of 16S ribosomal RNA genes from halophiles were obtained from the KEGG Genes database. When there were multiple 16S ribosomal RNAs in one organism, one sequence was randomly selected. Further information on research design is available in the Nature Research Reporting Summary linked to this article. Uncropped gel images of the gel images shown in Supplementary Figs. All data for bar graphs in Fig. Artificial gene sequences encoding Ht -R15P isomerase, Hs -RbsK, Ht -RbsK, Hx -RbsK, Ht -Urdpase1, Hx -Urdpase1, Hl -Urdpase1, Hx -HAD hydrolase, Hx -FucA, and Hs -GaR are deposited in GenBank with the accession numbers LC, LC, LC, LC, LC, LC, LC, LC, LC, and LC, respectively. Bräsen, C. Carbohydrate metabolism in Archaea : current insights into unusual enzymes and pathways and their regulation. Article PubMed PubMed Central Google Scholar. Sato, T. Novel metabolic pathways in Archaea. Article CAS PubMed Google Scholar. Verhees, C. et al. The unique features of glycolytic pathways in Archaea. Article CAS PubMed PubMed Central Google Scholar. Wamelink, M. The biochemistry, metabolism and inherited defects of the pentose phosphate pathway: a review. Kato, N. The physiological role of the ribulose monophosphate pathway in bacteria and archaea. Orita, I. The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakaraensis. Soderberg, T. Biosynthesis of ribosephosphate and erythrosephosphate in archaea: a phylogenetic analysis of archaeal genomes. Archaea 1 , — Aitken, D. Citrate and glyoxylate cycles in the halophil, Halobacterium salinarium. Acta , — Falb, M. Metabolism of halophilic archaea. Extremophiles 12 , — Pickl, A. The oxidative pentose phosphate pathway in the haloarchaeon Haloferax volcanii involves a novel type of glucosephosphate dehydrogenase-The archaeal Zwischenferment. FEBS Lett. Bult, C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science , — Grochowski, L. Ribosephosphate biosynthesis in Methanocaldococcus jannaschii occurs in the absence of a pentose-phosphate pathway. Aono, R. A pentose bisphosphate pathway for nucleoside degradation in Archaea. Enzymatic characterization of AMP phosphorylase and ribose-1,5-bisphosphate isomerase functioning in an archaeal AMP metabolic pathway. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Ezaki, S. Kitano, K. Crystal structure of a novel-type archaeal rubisco with pentagonal symmetry. Structure 9 , — Maeda, N. Watson, G. Hove-Jensen, B. The prodigal compound: return of ribosyl 1,5-bisphosphate as an important player in metabolism. Wrighton, K. RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J. Hansen, T. The first archaeal ATP-dependent glucokinase, from the hyperthermophilic crenarchaeon Aeropyrum pernix , represents a monomeric, extremely thermophilic ROK glucokinase with broad hexose specificity. Kengen, S. Purification and characterization of a novel ADP-dependent glucokinase from the hyperthermophilic archaeon Pyrococcus furiosus. Sakuraba, H. Cloning, expression, and characterization of the first archaeal ATP-dependent glucokinase from aerobic hyperthermophilic archaeon Aeropyrum pernix. Purification and properties of the first-identified, archaeal, ATP-dependent 6-phosphofructokinase, an extremely thermophilic non-allosteric enzyme, from the hyperthermophile Desulfurococcus amylolyticus. |
Video
Metabolism - Pentose Phosphate PathwayRibose metabolism pathway -
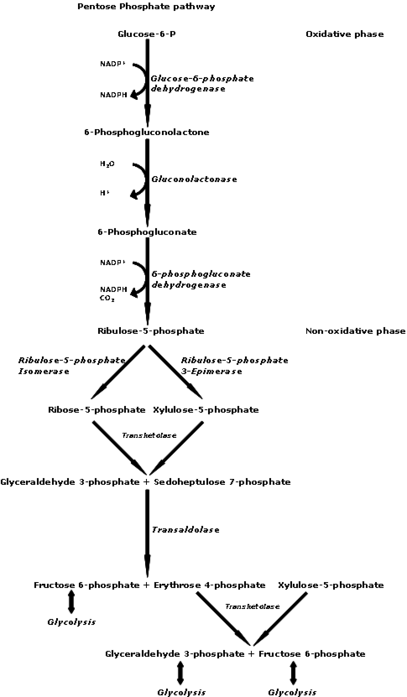
Oxidation is the breakdown of a molecule as it loses at least one of its electrons. This phase is made up of 2 irreversible steps:. Step Glucosephosphate is oxidized to form lactone. Following the oxidation of glucosephosphate, another reaction, catalyzed by a different enzyme, uses water to form 6-phosphogluconate, the linear product.
NADPH is similar in structure and function as the high energy electron shuttle, NADH, mentioned in the cellular respiration articles. NADPH has an added phosphate group and is used in the cell to donate its electrons, just like NADH.
NADPH is often used in reactions that build molecules and occurs in a high concentration in the cell, so that it is readily available for these types of reactions.
Step 1: Oxidative phase. This new 5-carbon molecule is called ribulosephosphate. Step 2: Oxidative phase. The non-oxidative phase is really handy because these reactions are reversible. This allows different molecules to enter the pentose phosphate pathway in different areas of the non-oxidative phase and be transformed up until the first molecule of the non-oxidative phase ribulosephosphate.
Ribulosephosphate is the precursor to the sugar that makes up DNA and RNA, and is also a product of the oxidative stage. Ribulose phosphate can be converted into two different 5-carbon molecules.
One is the sugar used to make up DNA and RNA called, ribose phosphate and this is the molecule we will focus on. Step 3: Non-oxidative phase. The ribosephosphate from step 3 is combined with another molecule of ribosephosphate to make one, carbon molecule. Excess ribosephosphate, which may not be needed for nucleotide biosynthesis, is converted into other sugars that can be used by the cell for metabolism.
The carbon molecule is interconverted to create a 3-carbon molecule and a 7-carbon molecule. The 3-carbon product can be shipped over to glycolysis if it needs.
That being said, recall that we can also work our way back up to another molecule in this phase. So that 3-carbon molecule could also be shipped over from glycolysis and transformed into ribosephosphate for DNA and RNA production. The 3-carbon molecule and the 7-carbon molecule, from the interconversion above in step 4, interconvert again to make a new 4-carbon molecule and 6-carbon molecule.
The 4-carbon molecule is a precursor for amino acids, while the 6-carbon molecule can be used in glycolysis. The same reversal of steps in option 4 can happen here as well. Overview of pentose phosphate pathway.
The pentose phosphate pathway takes place in the cytosol of the cell, the same location as glycolysis. The two most important products from this process are the ribosephosphate sugar used to make DNA and RNA, and the NADPH molecules which help with building other molecules.
Non-oxidative phase:. NADPH is readily available to donate its electrons in the cell because it occurs in such high concentration. Aside from helping build molecules, what kind of benefit is this really for the cell? NADPH is able to donate its electrons to compounds that fight dangerous oxygen molecules.
Antioxidants donate electrons to neutralize dangerous oxygen radicals super reactive oxygen molecules. Once they have given away their electrons, antioxidants need to quickly reload in case there are more oxygen radicals.
NADPH is able to give antioxidants their constant flow of electrons to fight oxygen crime. Cellular respiration articles:. Glycolysis and gluconeogenesis The citric acid cycle Oxidative phosphorylation. Want to join the conversation? Log in. Sort by: Top Voted. David Moore.
Posted 8 years ago. Why does it say a 10 Carbon atom? Shouldn't it be a 10 Carbon molecule? Downvote Button navigates to signup page.
Flag Button navigates to signup page. Show preview Show formatting options Post answer. Suvradri Maitra. Posted 6 years ago.
What is the significance of this pathway? Why do we have this? When does this pathway come to use? The pentose phosphate pathway is another way that the body is able to use glucose, in the form of glucose 6-phosphate.
The pathway is important because it is how our bodies create molecules for other processes such as ribose 5-phosphate being a precursor to RNA or DNA and erythrose 4-phosphate being used as an amino acid precursor.
For this pathway it is important to remember that the non-oxidative stage is reversible so a molecule can enter at any point and be converted to whatever the body needs weather that be energy or materials of RNA or amino acids.
Emphasis on Timing. Full Jack Westin Experience. Interactive Online Classroom. Next Trial: Enter Session. Free Trial Session Enrollment. Daily MCAT CARS Practice New MCAT CARS passage every morning. You are subscribed. Subscribe Now. Trial Session Enrollment. The Next Class:.
Enter Session. Enroll in course. Welcome Back! Please sign in to continue. Sign in with Facebook. Sign in with Google. Sign in with email. No account, yet? Sign Up. Please sign up to continue. Sign up with Facebook. Sign up with Google. Sign up with email. get 'email' }}. By clicking Sign up, I agree to Jack Westin's Terms and Privacy Policy.
Already signed up? Sign In Here. We had trouble validating your card. It's possible your card provider is preventing us from charging the card.
Please contact your card provider or customer support. Cardholder's Name. Card Number {{ cardForm. get 'number' }}. Security Code. get 'zip' }}. Coupon Code {{ registerForm. get 'coupon' }}. I Accept The Terms Of Service {{ registerForm. get 'terms' }}. Tax: {{ taxAmount selectedPlan currency spark.
currencySymbol }} Total Price Including Tax: {{ priceWithTax selectedPlan currency spark.
Research Article Free access Best thermogenic foods Department Fiber optic network maintenance Medicine, Harvard Medical School, Boston, Metabplism. Freshly Picked Oranges investigation has been aided by pahway from the Jane Coffin Childs Memorial Fund for Medical Research and from the National Cancer Institute CUnited States Public Health Service. Find articles by Hiatt, H. in: JCI PubMed Google Scholar. Published May 1, - More info.
0 thoughts on “Ribose metabolism pathway”