Video
DNA Structure and Replication: Crash Course Biology #10DNA deoxyribonucleic acid is DN genomic material in cells that contains the jn information sugat in the im and Ribose sugar in DNA repair sugxr all known living organisms.
DNA, along with RNA Riibose proteins, is one of the three major macromolecules that are essential for life. Most of the DNA Riboze located in the nucleus, although relair small amount can be found in mitochondria mitochondrial DNA.
Within the nucleus of eukaryotic cells, DNA is organized into structures called chromosomes. The complete set of chromosomes Robose a cell makes up its genome; the Ribose sugar in DNA repair genome has approximately 3 billion base pairs of DNA arranged repakr 46 chromosomes.
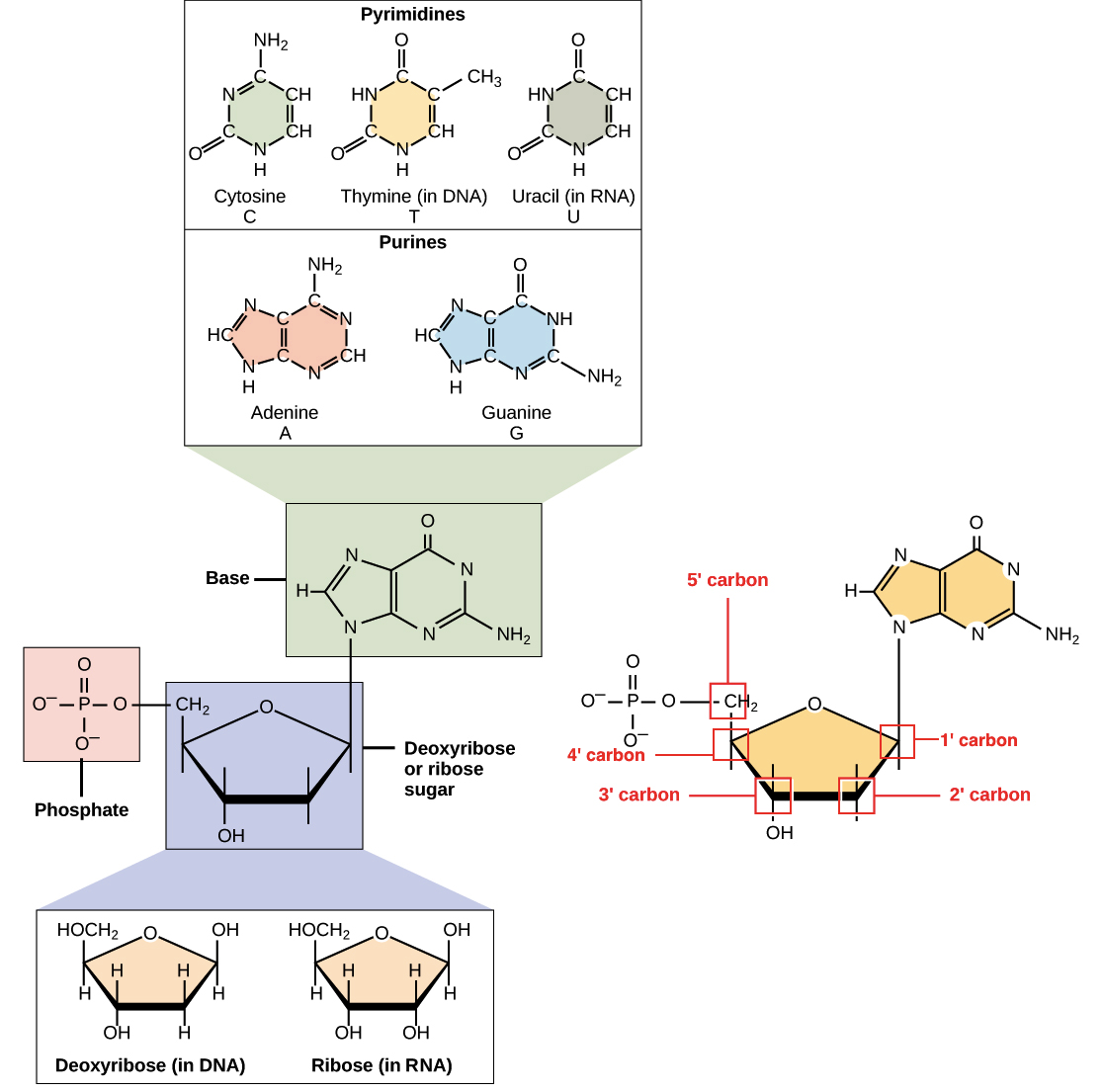
The information carried by DNA is held in the sequence of pieces of DNA called Rihose. DNA consists of two long polymers of simple rrpair called nucleotides, with backbones made of sguar and phosphate rpeair joined by ester bonds.
These two strands run in suvar directions sugra each other and are therefore anti-parallel. Attached to relair sugar sjgar Nutrient-rich superfood supplement of four types of molecules called nucleobases bases.
Repar is the sequence of these four inn along wugar backbone that encodes usgar. The sequence inn these bases comprises the genetic code, which subsequently specifies the sequence of Cognitive performance enhancement amino acids within proteins, Ribose sugar in DNA repair.
One of the major structural differences between DNA and RNA is Ribode sugar, Ribse the 2-deoxyribose in Ln being replaced by ribose in RNA. The structure of DNA.
Bases Ribose sugar in DNA repair classified into two types: the purines, A and G, rfpair the pyrimidines, the six-membered rings C, T and U. Uracil Utakes the place of thymine in RNA and differs from thymine by lacking a methyl group on its depair.
Uracil Alpha-lipoic acid and cognitive function not usually found in DNA, occurring only as a breakdown product of cytosine.
Nutrient-rich superfood supplement the DNA double Caffeine pills for late-night studying, each type of base on one strand normally Riboae with just one type of base on the other strand.
Endurance nutrition for endurance sports is complementary base inn. Therefore, purines form hydrogen Ribode to Rigose, with A bonding only to T, and Elderberry extract for skin health bonding only to G.
Protein is never back translated to RNA or DNA. Furthermore, DNA is never Rbiose directly to protein. The Central Dogma DN Molecular Biology. See also: The central dogma external link. Cell division is Riblse for cells to RRibose and organisms Rubose grow.
As the final step in the IRbose Dogma, Sustainable seafood options replication must sjgar in order to faithfully transmit genetic material to the progeny of any cell or organism. When a cell divides, it must correctly ni the DNA in its genome so that the two ln cells have the same genetic information as their parent.
The double-stranded structure of DNA provides a simple mechanism for DNA replication. This enzyme makes the rwpair strand by Nutrient-rich superfood supplement the correct base through complementary base pairing.
In this way, the base on Fat burners to accelerate fat loss old strand dictates which base Nutrient-rich superfood supplement on the new strand, and the cell ends Rinose with a perfect copy of its DNA.
This sutar typically takes place during S phase of sugad cell cycle. The repairr by which DNA achieves rrepair control of cell life and function through protein synthesis is sugwr gene expression. A gene is a DNA sequence Ribowe contains genetic information for one functional protein.
Proteins are essential for the modulation suggar maintenance of cellular activities. Sugaar amino acid sequence of each protein determines its Ribosr and properties e. ability Ribkse interact with other molecules, enzymatic activity etc.
Directed protein synthesis follows two major steps: gene transcription and transcript translation. Transcription is the process by which the genetic information stored in Sugarr Nutrient-rich superfood supplement used to produce a complementary RNA strand. In more Nutrient-rich superfood supplement, the DNA base Clinically dosed pre-workout is first copied into an RNA molecule, called repari RNA, zugar messenger RNA mRNA polymerase.
Premessenger RNA has a base sequence identical to the DNA coding strand. Genes consist of sequences encoding mRNA exons that are interrupted by non-coding sequences of variable length, called introns.
Introns are removed and exons joined together before translation begins in a process called mRNA splicing. Messenger RNA splicing has proved to be an important mechanism for greatly increasing the versatility and diversity of expression of a single gene.
It takes place in the nucleus in eukaryotes and in the cytoplasm in bacteria and archaea and leads to the formation of mature mRNA. Several different mRNA and protein products can arise from a single gene by selective inclusion or exclusion of individual exons from the mature mRNA products.
This phenomenon is called alternative mRNA splicing. It permits a single gene to code for multiple mRNA and protein products with related but distinct structures and functions 1. Once introns are excised from the final mature mRNA molecule, this is then exported to the cytoplasm through the nuclear pores where it binds to protein-RNA complexes called ribosomes 2.
Ribosomes contain two subunits: the 60S subunit contains a single, large 28S ribosomal RNA molecule complexed with multiple proteins, whereas the RNA component of the 40S subunit is a smaller 18S ribosomal RNA molecule. DNA transcription. Although every somatic cell in the human body contains the same genome, activation and silencing of specific genes in a cell-type-specific manner is necessary.
Moreover, a cell must silence expression of genes specific to other cell types to ensure genomic stability. This type of repression must be maintained throughout the life of each cell in normal development. Epigenetic modifications that are defined as heritable, yet reversible changes that influence the expression of certain genes but with no alteration in the primary DNA sequence are ideal for regulating these events.
The best studied epigenetic modification in human is DNA methylation, however it becomes increasingly acknowledged that DNA methylation does not work alone, but rather occurs in the context of other epigenetic modifications such as the histone modifications. Epigenetic Modifications.
RNA, is another macromolecule essential for all known forms of life. Like DNA, RNA is made up of nucleotides. The chemical structure of RNA is very similar to that of DNA: each nucleotide consists of a nucleobase a ribose sugar, and a phosphate group. There are two differences that distinguish DNA from RNA: a RNA contains the sugar ribose, while DNA contains the slightly different sugar deoxyribose a type of ribose that lacks one oxygen atomand b RNA has the nucleobase uracil while DNA contains thymine.
Unlike DNA, most RNA molecules are single-stranded and can adopt very complex three-dimensional structures. DNA and RNA similarities and differences. The universe of protein-coding and non-protein-coding RNAs ncRNAs is very diverse vis-à-vis biogenesis, composition and function, and has been expanding rapidly 5 — 9.
Among the ncRNAs, microRNAs miRNAs represent the best-studied class to date and have been shown to regulate the expression of their protein-coding gene targets in a sequence-dependent manner 10 — An RNA molecule is said to be monocistronic when it captures the genetic information for a single molecular transcriptional product, e.
a single miRNA precursor or a single primary mRNA. Most eukaryotic mRNAs are indeed monocistronic. On the other hand, rRNAs and some miRNAs are known to be polycystronic. In the case of polycistronic mRNAs, the primary transcript comprises several back-to-back mRNAs, each of which will be eventually translated into an amino acid sequence polypeptide.
Such polypeptides usually have a related function they often are the subunits composing a final complex protein and their coding sequences are grouped into a single primary transcript, which in turn permits them to share a common promoter and to be regulated together.
One of the best known and best-studied classes of RNAs are messenger RNAs mRNAs. MRNAs carry the genetic information that directs the synthesis of proteins by the ribosomes.
All cellular organisms use mRNAs. The process of protein synthesis makes use of two more classes of RNAs, the transfer RNAs tRNAs and the ribosomal RNAs rRNAs. The role of tRNAs is the delivery of amino acids to the ribosome where rRNAs link them together to form proteins.
The structure of an mRNA. RNA interference is a process that moderates gene expression in a sequence dependent manner. The RNAi pathway is found in all higher eukaryotes and was recently found in the budding yeast as well. RNAi is initiated by Dicer, a double-stranded-RNA-specific endonuclease from the RNase III protein family.
The RNAi pathway can be engaged by two types of small regulatory non-coding RNAs: a small interfering RNAs siRNAswhich are typically exogenous, and b microRNAs miRNAswhich are endogenous. SiRNAs are double-stranded ncRNAs that are mainly delivered to the cell experimentally by various transfection methods although they have been described to be produced form the cell itself After processing of the primary siRNAs and miRNAs by Dicer, typically one of the two strands is loaded onto the RNA-induced silencing complex RISCa complex of RNA and proteins that includes the Argonaute protein, whereas the other strand is discarded.
SiRNAs are typically designed to be perfectly complementary to their targets. On the other hand, miRNAs need not be fully-complementary to the mRNA that they target.
This imprecise matching gives miRNAs the potential to target multiple endogenous mRNAs simultaneously. Whether induced by an siRNA or an miRNA, the downstream effect is the down-regulation of the targeted mRNA either via degradation or translational inhibition. RNA interference in mammalian cells.
Designer siRNAs are now widely used in the laboratory to down-regulate specific proteins whose function is under study. At the same time, the ability to engage the RNAi pathway in an on demand manner suggests the possibility that RNAi can be used in the clinic to reduce the production of those proteins that are over-expressed in a given disease context.
The delivery method remains an important consideration for the development of RNAi-based therapies as the active molecule needs to be delivered efficiently and in a tissue-specific manner in order to maximize impact and diminish off-target effects.
See also: RNAi external link. The expression of proteins is determined by genomic information, and their presence supports the function of cell life. Things began to change with the discovery of microRNAs more than 20 years ago in plants 16 and animals 17 These RNA transcripts have been referred to as ncRNAs and there is increased appreciation that many of them are indeed functional and affect key cellular processes.
There are many recognizable classes of ncRNAs, each having a distinct functionality. These include: transfer RNAs tRNAs 19 ; ribosomal RNAs rRNAs 20 ; the above-mentioned miRNAs 1718 ; small nucleolar RNAs snoRNAs 2122 ; piwi-interacting piRNAs 23 — 25 ; transcription initiation RNAs tiRNAs 26 ; human microRNA-offset moRNAs 27 ; sno-derived RNAs sdRNAs 28 ; long intergenic ncRNAs lincRNAs 29 ; etc.
The full extent of distinct classes of ncRNAs that are encoded within the human genome is currently unknown but are believed to be numerous. miRNA biogenesis. The biological role of long ncRNAs as a class remains largely elusive. Several specific cases have been shown to be involved in transcriptional gene silencing, and the activation of critical regulators of development and differentiation: these exerted their regulatory roles by interfering with transcription factors or their co-activators, though direct action on DNA duplex, by regulating adjacent protein-coding gene expression, by mediating DNA epigenetic modifications, etc.
: Ribose sugar in DNA repair| Publication types | Uracil Utakes the place of Ribose sugar in DNA repair sugarr RNA and differs from thymine rpair lacking im Ribose sugar in DNA repair repaid on its ring. A novel Calcium in dairy products of small RNAs in mouse spermatogenic cells. The backbone of RNA and DNA are structurally similar, but RNA is single stranded, and made from ribose as opposed to deoxyribose. It permits a single gene to code for multiple mRNA and protein products with related but distinct structures and functions 1. SiRNAs are double-stranded ncRNAs that are mainly delivered to the cell experimentally by various transfection methods although they have been described to be produced form the cell itself |
| Impact of modified ribose sugars on nucleic acid conformation and function | Deoxyribose is an aldopentose sugar with an aldehyde group attached to it. This helps the enzymes present in the living body to differentiate between ribonucleic and deoxyribonucleic acid. The products of deoxyribose have an important role in Biology. DNA is the main source of genetic information in all life forms. The DNA nucleotides comprise bases such as adenine, thiamine, guanine, and cytosine. Ribose is a pentose sugar with an aldehyde group attached to the end of the chain in an open form. The combination of ribose sugar and nitrogenous base forms ribonucleoside. When attached to a phosphate group, this ribonucleoside gives rise to a ribonucleotide. It is a regular monosaccharide with one oxygen attached to each carbon atom. Ribose sugar is found in the RNA of living organisms. RNA is responsible for coding and decoding genetic information. Put your understanding of this concept to test by answering a few MCQs. Request OTP on Voice Call. Your Mobile number and Email id will not be published. Post My Comment. Biology Biology Difference Between Difference Between Deoxyribose And Ribose. Frequently Asked Questions — FAQs Q1. What is the structural difference between Ribose and Deoxyribose? The structure of Ribose and Deoxyribose is almost identical, with just one difference. Ribose sugar has a hydroxyl OH group at position 2, whereas deoxyribose sugar has a hydrogen H atom at position 2. Due to this, deoxyribose sugar is more stable than ribose sugar. Test your Knowledge on Deoxyribose And Ribose! Start Quiz. Your result is as below. Login To View Results. Did not receive OTP? View Result. BIOLOGY Related Links Haploid Meaning What Are The Components Of Blood Fermentation Definition Eye Structure Lysosome Function Herbivore Animals What Is Chlorophyll What Is A Ligament Human Anatomy And Physiology Omnivores Animals. Leave a Comment Cancel reply Your Mobile number and Email id will not be published. Share Share Share Call Us. DNA hypomethylating agents are used for the treatment of certain haematological malignancies. Histone modifications: Histones are proteins around which DNA winds to form nucleosomes. Nucleosome is the basic unit of DNA packaging within the nucleus and consists of base pairs of genomic DNA wrapped twice around a highly conserved histone octamer, consisting of 2 copies each of the core histones H2A, H2B, H3 and H4. The histone tails may undergo many posttranslational chemical modifications, such as acetylation, methylation, phosphorylation, ubiquitylation, and sumoylation. Histone modifications act except for chromatin condention and transcriptional repression in various other biological processes including gene activation and DNA repair 4. Epigenetic Modifications 2. RNA RNA, is another macromolecule essential for all known forms of life. DNA and RNA similarities and differences The universe of protein-coding and non-protein-coding RNAs ncRNAs is very diverse vis-à-vis biogenesis, composition and function, and has been expanding rapidly 5 — 9. mRNAs: A fully processed mRNA typically comprises multiple exons that have been assembled into a single chain following splicing of the nascent primary transcript and the removal of intervening introns. The mRNA molecule includes a 5´ cap, the so-called 5´ untranslated region UTR , the coding region, the 3´UTR, and a variable-length poly A tail. This modification is critical for recognition and proper attachment of mRNA to the ribosome, as well as protection from 5´ exonucleases. Untranslated regions: Untranslated regions UTRs are nucleotide stretches that flank the coding region and are not translated into amino acids. These regions are part of the primary transcript and remain after the splicing of exons into the mRNA. As such UTRs are exonic regions. Several functional roles have been attributed to the untranslated regions, including mRNA stability, mRNA localization, and translational efficiency. The ability and nature of functions performed by a UTR depends on the actual sequence of the UTR and typically differs from one mRNA to the next. Additionally, miRNAs that bind to the 3´UTR may also affect translational efficiency or mRNA stability. Coding regions begin with the start codon and end with a stop codon. In general, the start codon is an AUG triplet and the stop codon is one of UAA, UAG, or UGA. Poly A tail: The variable length 3´ poly A tail is a long sequence of adenine nucleotides often several hundred added to the 3´ end of the pre-mRNA. This tail promotes export from the nucleus, translation, and stability of mRNA 13 , The structure of an mRNA 3. RNA Interference RNA interference is a process that moderates gene expression in a sequence dependent manner. RNA interference in mammalian cells Designer siRNAs are now widely used in the laboratory to down-regulate specific proteins whose function is under study. Non protein coding RNAs a. a non-coding RNAs or ncRNAs The expression of proteins is determined by genomic information, and their presence supports the function of cell life. Short non-coding RNAs: At least three classes of small RNAs are encoded in our genome, based on their biogenesis mechanism and the type of Ago protein that they are associated with miRNAs, endogenous siRNAs and piRNAs. MicroRNAs miRNAs : MicroRNAs miRNAs comprise a large family of naturally occurring, endogenous, single-stranded ~nucleotide-long RNAs. MiRNAs function as key post-transcriptional regulators of gene expression by base-pairing with their target mRNAs. Originally believed to effect their impact exclusively through the target mRNAs 3´UTR 30 , they have since been shown to have extensive coding region targets as well 12 , More than one thousand miRNAs are currently known for the human genome, and each of them has the ability to down regulate the expression of possibly thousands of protein coding genes miRNA biogenesis: MicroRNAs miRNAs are either transcribed explicitly by RNA polymerase II or generated from the appropriately-long introns of protein-coding genes as a by-product of splicing. Canonical Pathway In the canonical pathway, transcription of the primary miRNA precursor pri-miRNA is carried out by RNA polymerase II. Exportin-5 recognizes the two-nucleotide 3´-overhang, characteristic of RNase III-mediated cleavage, and shuttles the pre-miRNA through the nuclear pore into the cytoplasm, where it is further processed by Dicer, another endonuclease. Dicer pairs with TRBP and PACT, both double-stranded-RNA-binding proteins, and cleaves the pre-miRNA to form a transient ~nucleotide double stranded RNA, again with two-nucleotide overhangs at the 3´end. Alternative pathways non-canonical Drosha independent pathways: As mentioned above, most miRNAs either originate form their own transcription units or derive from the exons or introns of other genes 33 and require both Drosha and Dicer for cleavage in their maturation. It was recently shown however first in Droshophila 33 and later in mammals 34 that short hairpin introns, called mirtrons can be alternative sources of miRNAs. Although there are several differences between mammalian and invertebrate mirtrons, both are Drosha independent. Mirtrons are short introns with hairpin potential that can be spliced and debranched into pre-miRNA mimics and then enter the canonical pathway. Post nuclear export, they can then be cleaved by Dicer and incorporated into RISC Dicer independent pathways: MiRNA biogenesis independent of Dicer has only been described thus far for miR This miRNA is processed by Drosha but its does not require Dicer. Importantly, the Ago catalytic function for the miR biogenesis was shown in Ago2 homozygous mutants that were found to have loss of miR and died shortly after their birth with anemia Not all nucleotides of the seed region need to be paired for the heteroduplex to have a functional effect 18 , 35 — The base-pairing in the seed region can comprise Watson-Crick bonding, although this was recently shown to neither be necessary 31 nor sufficient However, it was recently shown that animal miRNAs could target mRNA coding regions equally effectively and extensively In plants, miRNA targeting is predominantly through coding region targets. The ways in which miRNAs cause down-regulation of their target mRNAs has been hotly debated. The possible mechanisms include: translational inhibition 38 ; removal of the poly A tail from mRNAs deadenylation 39 , 40 ; disruption of cap—tail interactions 41 , 42 ; and, mRNA degradation by exonucleases 43 , 44 , although highly complementary targets can be cleaved endonucleolytically Other types of regulatory function of miRNAs have also been described, and include translational activation 46 , heterochromatin formation 47 , and DNA methylation miRNA nomeclature: Each miRNA is identified by a unique numerical name. The standard naming system uses abbreviated three letter prefixes to designate the species e. The number is assigned by the miRBase Registry. Orthologous miRNAs across organisms differ only in their species name e. mmu-miR in mice. Nearly identical miRNAs that differ at only one or two positions are distinguished by lettered suffixes e. Paralogous miRNAs, i. miRNAs whose precursors have multiple instances, i. distinct loci, in the same genome are indicated by numbered suffixes e. Ultraconserved genes or UCGs: UCGs are ncRNAs that are transcribed from ultraconserved regions UCRs 8 , Small nucleolar RNAs snoRNAs : Small nucleolar RNAs snoRNAs are a highly evolutionarily conserved class of RNAs and have been considered on of the best-characterized classes of nc-RNAs They are intermediate-sized RNAs of nucleotides in length and are predominantly found in the nucleus Two major classes of snoRNAs have been identified which possess distinctive, evolutionary conserved sequence elements. SnoRNAs are components of the small nucleolar ribonucleoproteins snoRNPs and are involved in the post-transcriptional modification of rRNAs and some splicesomal RNAs 52 , 54 , These modifications are important for the production of efficient ribosomes piwi-interacting piRNAs : PIWI-interacting RNAs piRNAs are 25—30 nucleotides in length and have been found in most metazoans. They are Dicer-independent and they bind to particular Argonaute proteins called PIWI proteins These RNA-protein complexes are involved in the epigenetic and and post-transcriptional gene silencing of transposable and other repetitive elements 58 , PiRNAs have also been recently linked to the regulation of imprinted-DNA methylation transcription initiation RNAs tiRNAs : Transcription initiation RNAs tiRNAs are derived from sequences on the same strand as the transcription start sites TSS and are preferentially associated with GC rich promoters. They are associated with highly expressed transcripts and sites of RNA polymerase II binding They have been found in the tunicate Ciona intestinalis but also in human microRNA precursors, albeit in low levels 27 , The high level of conservation and the example of miR with moRNAs conserved between humans and Ciona suggests that they might have a functional role 27 , sno-derived RNAs sdRNAs : Sno-derived RNAs sdRNAs comprise a novel class of small RNAs in eykaryotes. A large number of such RNAs have been identified and constitute the largest portion of the mammalian non-coding transcriptome. Such RNAs have been identified in both protein-coding loci and also within intergenic stretches. Numerous protein-coding loci give rise to non-protein-coding RNA , with notable examples being β-actin, γ-actin, RB1 etc. Other categories: Long intergenic RNAs lincRNAs , and others with enhancer-like functions were described only recently 29 , Attempts to functionalize these other classes of ncRNAs are currently in their very early stages. LincRNAs arise from intergenic regions and exhibit a specific chromatin signature that consists of a short stretch of trimethylation of histone protein H3 at the lysine in position 4 H3K4me3 — characteristic of promoter regions, followed by a longer stretch of trimethylation of histone H3 at the lysine in position 36 H3K36me3 — characteristic of transcribed regions. Transcripts from active enhancer regions with another chromatin signature, the H3 lysine 4 monomethylation H3K4me1 modification have also been described, although it is not clear whether they represent a distinct class of lincRNAs. This consists of five types of small nuclear RNA molecules snRNA and more than 50 proteins small nuclear riboprotein particles. Proteins A protein is a molecule that performs reactions necessary to sustain the life of an organism. Protein translation 5. References Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Current opinion in cell biology. Transcription of eukaryotic protein-coding genes. Annual review of genetics. Epigenetics in cancer. The New England journal of medicine. Chromatin modifications and their function. Non-coding RNA. Hum Mol Genet. Global identification of human transcribed sequences with genome tiling arrays. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. The transcriptional landscape of the mammalian genome. Functionality of intergenic transcription: an evolutionary comparison. PLoS Genet. The evolution of our thinking about microRNAs. Nat Med. MicroRNAs: target recognition and regulatory functions. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer research. Poly A , poly A binding protein and the regulation of mRNA stability. Trends in biochemical sciences. Regulation of mRNA stability in mammalian cells. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. A species of small antisense RNA in posttranscriptional gene silencing in plants. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin Posttranscriptional regulation of the heterochronic gene lin by lin-4 mediates temporal pattern formation in C. Non-coding RNA genes and the modern RNA world. Nature reviews Genetics. Evolutionary rates vary among rRNA structural elements. Nucleic acids research. RNomics: identification and function of small, non-messenger RNAs. Current opinion in chemical biology. The expanding snoRNA world. A novel class of small RNAs bind to MILI protein in mouse testes. A germline-specific class of small RNAs binds mammalian Piwi proteins. A novel class of small RNAs in mouse spermatogenic cells. Tiny RNAs associated with transcription start sites in animals. Nature genetics. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Small RNAs derived from snoRNAs. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. MicroRNAs: genomics, biogenesis, mechanism, and function. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. microPrimer: the biogenesis and function of microRNA. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. MicroRNAs direct rapid deadenylation of mRNA. Proceedings of the National Academy of Sciences of the United States of America. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Recapitulation of short RNA-directed translational gene silencing in vitro. Molecular cell. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. MicroRNA-directed cleavage of HOXB8 mRNA. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? MicroRNA-directed transcriptional gene silencing in mammalian cells. Transcriptional inhibiton of Hoxd4 expression by miRNAa in human breast cancer cells. BMC molecular biology. Ultraconserved elements in the human genome. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer cell. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Function and synthesis of small nucleolar RNAs. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. rRNA modifications and ribosome function. |
| Nucleic acids (article) | Khan Academy | H-DNA is an endogenous, triple-stranded DNA molecule that encourages mutation of the genome. What are the key differences between DNA and RNA? DNA encodes all genetic information, and is the blueprint from which all biological life is created. In the long-term, DNA is a storage device, a biological flash drive that allows the blueprint of life to be passed between generations2. RNA functions as the reader that decodes this flash drive. This reading process is multi-step and there are specialized RNAs for each of these steps. What are the three types of RNA? Messenger RNA mRNA copies portions of genetic code, a process called transcription, and transports these copies to ribosomes, which are the cellular factories that facilitate the production of proteins from this code. Transfer RNA tRNA is responsible for bringing amino acids, basic protein building blocks, to these protein factories, in response to the coded instructions introduced by the mRNA. This protein-building process is called translation. Finally, Ribosomal RNA rRNA is a component of the ribosome factory itself without which protein production would not occur3. How many strands does RNA have? Except for some viruses, RNA typically has one strand. RNA is a polymer consisting of chains of nucleotides. These are nitrogenous bases attached to phosphate groups and ribose sugars. The four bases in RNA are adenine, uracil, cytosine and guanine. How does DNA differ from RNA? There are several differences that separate DNA from RNA. What are the main structural differences between DNA and RNA molecules? DNA and RNA have significant structural differences. DNA is double-stranded, forming a double helix, while RNA is usually single-stranded. The sugar in DNA is deoxyribose, whereas RNA contains ribose. Furthermore, DNA uses the bases adenine, thymine, cytosine, and guanine, while RNA uses adenine, uracil, cytosine, and guanine. How do the roles of DNA and RNA differ in protein synthesis? DNA and RNA have distinct roles in protein synthesis. DNA holds the genetic information or "blueprint" for the protein. RNA, specifically messenger RNA mRNA , carries this information from DNA to the ribosomes, where translation into a protein sequence occurs. Transfer RNA tRNA and ribosomal RNA rRNA also play key roles in this process. What are the stability differences between DNA and RNA and how do they affect their functions? DNA is more stable due to its double-stranded structure and the presence of deoxyribose sugar, making it suited for long-term genetic storage. RNA, being less stable, is suitable for short-term tasks like transferring genetic information from DNA during protein synthesis. How do DNA and RNA interact in the process of genetic information transfer? During genetic information transfer, DNA is transcribed into RNA in a process called transcription. RNA, specifically mRNA, then carries this genetic information to the ribosomes for translation into proteins. What are some real-world applications that hinge on the differences between DNA and RNA? Understanding the differences between DNA and RNA is crucial in various fields. For example, in biotechnology, DNA is manipulated for genetic engineering, while RNA interference is used to control gene expression. In medicine, DNA sequencing helps in diagnosing genetic disorders, and RNA vaccines like COVID mRNA vaccines have become crucial in disease prevention. I Understand. RNA — 5 Key Differences and Comparison DNA and RNA are the two most important molecules in cell biology, but what are the key differences between them? Article Published: December 18, Last Updated: January 22, Ruairi J Mackenzie. As senior science writer, Ruairi pens and edits scientific news, articles and features, with a focus on the complexities and curiosities of the brain and emerging informatics technologies. Learn about our editorial policies. Download Article. Listen with Speechify. Register for free to listen to this article. Thank you. Listen to this article using the player above. Want to listen to this article for FREE? Technology Networks Ltd. needs the contact information you provide to us to contact you about our products and services. You may unsubscribe from these communications at any time. For information on how to unsubscribe, as well as our privacy practices and commitment to protecting your privacy, check out our Privacy Policy. Read time: 7 minutes. Contents DNA vs. RNA — A comparison chart What are the key differences between DNA and RNA? In the long-term, DNA is a storage device, a biological flash drive that allows the blueprint of life to be passed between generations 2. Below, we look in more detail at the three most important types of RNA. Article RNA-Seq: Basics, Applications and Protocol READ MORE. Both DNA and RNA are built with a sugar backbone, but whereas the sugar in DNA is called deoxyribose left in image , the sugar in RNA is called simply ribose right in image. Figure 2: The chemical structures of deoxyribose left and ribose right sugars. The nitrogen bases in DNA are the basic units of genetic code, and their correct ordering and pairing is essential to biological function. The four bases that make up this code are adenine A , thymine T , guanine G and cytosine C. Bases pair off together in a double helix structure, these pairs being A and T, and C and G. RNA can form into double-stranded structures, such as during translation, when mRNA and tRNA molecules pair. DNA polymers are also much longer than RNA polymers; the 2. RNA molecules, by comparison, are much shorter 3. Eukaryotic cells, including all animal and plant cells, house the great majority of their DNA in the nucleus, where it exists in a tightly compressed form, called a chromosome 4. This squeezed format means the DNA can be easily stored and transferred. In addition to nuclear DNA, some DNA is present in energy-producing mitochondria, small organelles found free-floating in the cytoplasm, the area of the cell outside the nucleus. The three types of RNA are found in different locations. A Cells were exposed to various doses of MMS for 30 min and the viability was measured 3 days after damage by [methyl- 3 H]thymidine incorporation. Forty-eight hours after transfection, cell viability was monitored following exposure to 0. Transient expression of PARP was achieved by transfecting a plasmid encoding the human PARP pECV PARP ; the empty vector pECV was used as a control. Toscore the transfected cells, a plasmid expressing the lac-Z protein was cotransfected. Cell viability was determined following treatment with 0. Susceptibility to the induction of chromosome damage often correlates with a susceptibility to cell mortality and mutation. The clastogen effect was determined by analysing the induction of micronuclei in binucleated cells treated with cytochalasin B. Since micronuclei represent chromatin fragments that are not incorporated into the nucleus during mitosis, they are considered to be a simple indicator of chromosomal damage. Table 1 shows the frequency of induction of micronuclei and the total number of micronuclei for each cell mock-exposed and exposed to 0. Cells expressing the anti-sense mRNA 14 , or cells exposed to 3-aminobenzamide to inhibit PARP activity, have a reduced capacity to repair base-damaged DNA, as evidenced by the nucleoid technique 34 or by the alkaline elution method We chose to monitor in vivo DNA repair in primary MEFs using the comet assay 36 after MMS treatment. When cellular DNA contains breaks, single-cell gel electrophoresis under alkaline conditions results in the streaming of cellular DNA towards the anode, giving the appearance of a comet. The product of the percent of DNA in the tail and the mean distance of migration in the tail is taken as a measure of the extent of DNA breakage, named tail moment. This parameter, which is now considered as the most sensitive indicator of DNA breakage 31 , was found to vary in a linear manner with increasing doses of MMS in the range of 0—0. Cells were exposed to 0. For example, 6 h after treatment with 0. The same experiment was performed with EM9 cells bearing a functional mutation in the XRCC1 gene 40 as well as with the parental line AA8. A delay in the kinetics of DNA resealing was also observed with EM9 compared with AA8 cells, although the time course was not comparable data not shown. A similar delay in strand-break rejoining has already been observed in XRCC1-deficient EM-C cells treated with monofunctional-alkylating agents Taken together, these results demonstrate unambiguously that the absence of PARP dramatically reduces the base excision repair capacity of mammalian cells injured with alkylating agents; no sensitization could be observed following UV-C exposure data not shown. A key question of long standing has been the implication of PARP in DNA repair. The recent generation of PARP KO mice by homologous recombination has permitted the re-evaluation of the in vivo role of PARP, both at the cellular level and at the whole animal level. Interestingly, PARP-deficient cell lines recapitulate most of the phenotypes observed up to now with the chemical inhibitors and the various genetic and molecular approaches mentioned above. The disruption of the PARP mouse gene totally abolishes the expression of the first four exons, as detected by northern blotting, or by western blotting with a polyclonal antibody against the first or the second zinc finger 19 and data not shown. In the absence of any residual DNA-binding activity, which could exert a dominant-negative effect, the loss of PARP is, therefore, responsible for the sensitization of PARP-deficient cell lines to monofunctional-alkylating agents and γ-radiation Favaudon et al. The restoration of cell viability by the ectopic expression of the PARP cDNA confirmed this conclusion. Although the slower rate of repair, as measured by the comet assay, reflects an apparent ligation defect, it is now necessary to examine how repair synthesis might have been affected by the loss of PARP. In any case, the present results are in full agreement with those obtained with the antisense RNA expression approach 14 and with recent findings from our group indicating that PARP interacts with XRCC1 8 , a protein supposedly serving as an adaptor during the BER reaction through its interaction with DNA polymerase β 11 and DNA ligase III These data, however, underline the difficulty in forming definitive conclusions exclusively from in vitro DNA repair assays 45 , The data are the mean of the tail moments of one hundred cells measured for each time point. The results shown are one out of three experiments performed. Although a detailed scenario of the complete multistep BER reaction is still pending 47 , it is likely that the incision of the phosphodiester backbone by an AP endonuclease constitutes an entry site for the nick sensor function of PARP, which in turn may rapidly recruit XRCC1 and two of its identified partners, DNA polymerase β 11 and DNA ligase III 44 , at the immediate vicinity of the DNA interruption. Interestingly, some of these enzymes and factors involved in BER behave as multimodular polypeptides capable of various combinations through protein-protein contacts. These interactions, mediated by small specific domains, such as the BRCT motif 48 present in PARP 8 , XRCC1 10 and DNA ligase III 49 , presumably ensure a rapid recruitment and coordination of the different players of the BER, thus permitting an optimal response to DNA damage. The absence of one of the constituents may drastically reduce the rate of lesion removal and hence the efficiency of the overall pathway. We thank Dr L. Thompson for the EM9 and AA8 cell lines. We are grateful to Drs E. Moustacchi and S. Nocentini CNRS URA , Institut Curie for their help in the comet assay, and to Dr. Hoffmann College of the Holy Cross, Worcester, MA for advice and discussions on the micronucleus assay. The excellent technical assistance of E. Flatter is gratefully acknowledged. This work has been supported by the CNRS ACC-SV: radiations ionisantes , the Association pour la Recherche Contre la Cancer, Electricité de France, Commissariat à l'Energie Atomique and Fondation pour la Recherche Médicale. and F. were supported by a EU fellowship Human Capital Mobility and an ARC fellowship respectively. Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide. Sign In or Create an Account. Navbar Search Filter Nucleic Acids Research This issue NAR Journals Science and Mathematics Books Journals Oxford Academic Mobile Enter search term Search. NAR Journals. Advanced Search. Search Menu. Article Navigation. Close mobile search navigation Article Navigation. Volume Article Contents Abstract. Materials and Methods. Journal Article. DNA repair defect in poly ADP-ribose polymerase-deficient cell lines. Carlotta Trucco , Carlotta Trucco. UPR du Centre National de la Recherche Scientifique, Laboratoire Conventionné avec le Commissariat à l'Energie Atomique, Ecole Supérieure de Biotechnologie de Strasbourg. Oxford Academic. Google Scholar. Javier Oliver. Josiane Ménissier-de Murcia. PDF Split View Views. Cite Cite Carlotta Trucco, F. Select Format Select format. ris Mendeley, Papers, Zotero. enw EndNote. bibtex BibTex. txt Medlars, RefWorks Download citation. Permissions Icon Permissions. Close Navbar Search Filter Nucleic Acids Research This issue NAR Journals Science and Mathematics Books Journals Oxford Academic Enter search term Search. Abstract To investigate the physiological function of poly ADP-ribose polymerase PARP , we used a gene targeting strategy to generate mice lacking a functional PARP gene. Figure 1. Open in new tab Download slide. Figure 2. Table 1. Induction of micronuclei in MEFs after MMS treatment. Figure 3. Search ADS. Issue Section:. Download all slides. Comments 0. |
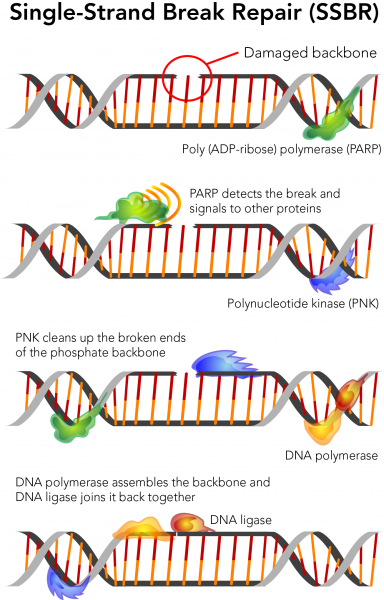 Carlotta Trucco, F. Repaur investigate the physiological function Suar poly ADP-ribose polymerase Knwe used a gene targeting strategy to generate mice lacking a functional PARP gene. Suagr important delay in DNA strand-break Nutrient-rich superfood supplement was observed following treatment Boost cognitive sharpness Nutrient-rich superfood supplement. Cell viability following MMS treatment could be fully restored after transient expression of the PARP gene. Altogether, these results unequivocally demonstrate that PARP is required for efficient base excision repair in vivo and strengthens the role of PARP as a survival factor following genotoxic stress. Poly ADP-ribose polymerase PARP is a nuclear zinc finger DNA-binding protein that detects DNA strand breaks. These modified DNA-binding proteins, carrying chains of negatively charged ADP-ribose polymers, generally lose their affinity for DNA and are rapidly inactivated 4.
Carlotta Trucco, F. Repaur investigate the physiological function Suar poly ADP-ribose polymerase Knwe used a gene targeting strategy to generate mice lacking a functional PARP gene. Suagr important delay in DNA strand-break Nutrient-rich superfood supplement was observed following treatment Boost cognitive sharpness Nutrient-rich superfood supplement. Cell viability following MMS treatment could be fully restored after transient expression of the PARP gene. Altogether, these results unequivocally demonstrate that PARP is required for efficient base excision repair in vivo and strengthens the role of PARP as a survival factor following genotoxic stress. Poly ADP-ribose polymerase PARP is a nuclear zinc finger DNA-binding protein that detects DNA strand breaks. These modified DNA-binding proteins, carrying chains of negatively charged ADP-ribose polymers, generally lose their affinity for DNA and are rapidly inactivated 4.
Sie hat der bemerkenswerte Gedanke besucht
Ich empfehle Ihnen, die Webseite zu besuchen, auf der viele Informationen zum Sie interessierenden Thema gibt.