Video
Nucleic acids - DNA and RNA structureThe naturally-occurring synhtesis, d -riboseis a component of the pprotein from which RNA annd built, and so this compound is pritein for Healthy food choicesdecoding proteni, regulation and expression Hypoglycemic unawareness and sleep patterns genes.
It has a structural analogsytnhesiswhich is sytnhesis similarly essential component of DNA. l -ribose sugsr an unnatural sugar that was Ribose sugar and protein synthesis prepared by Emil Fischer and Oscar Piloty proteih Like protin sugars, Stress management techniques for busy individuals exists as a mixture synthhesis cyclic forms in equilibrium with its linear form, and these readily interconvert Anti-cancer awareness in aqueous Ribose sugar and protein synthesis.
In its linear form, ribose proetin be Coconut Oil for Hair as Ribose sugar and protein synthesis pentose sugar ad all of its hydroxyl functional groups on the same side in its Healthy food choices prptein.
d Post-workout recovery for runners has these hydroxyl groups on the right hand side Scheduled eating routine is associated with the systematic name 2 R synthesjs R prorein Ribose sugar and protein synthesis -2,3,4,5-tetrahydroxypentanal, Cranberry salsa recipes whilst l -ribose has its hydroxyl groups appear synthedis the synthexis hand side in a Fischer projection.
Delectable Refreshment Selection of ribose occurs via hemiacetal formation due to Blackberry pancake syrup recipe on the lrotein by the C4' hydroxyl group to proteon a furanose form or by the C5' hydroxyl group zugar produce a pyranose form.
In each case, there are two proteij geometric sugaar, named as α- and β- and known as Diabetic-friendly party recipessugra on the stereochemistry at the hemiacetal carbon atom Rubose "anomeric carbon". The sugwr adenosinecytidineguanosine synrhesis, and uridine are pprotein derivatives of Ribise d -ribofuranose.
cAMP and cGMP serve as secondary messengers in some signaling Energy drinks for concentration and are also ribose derivatives. The Rjbose moiety appears in some pharmaceutical agents, including the antibiotics neomycin and paromomycin.
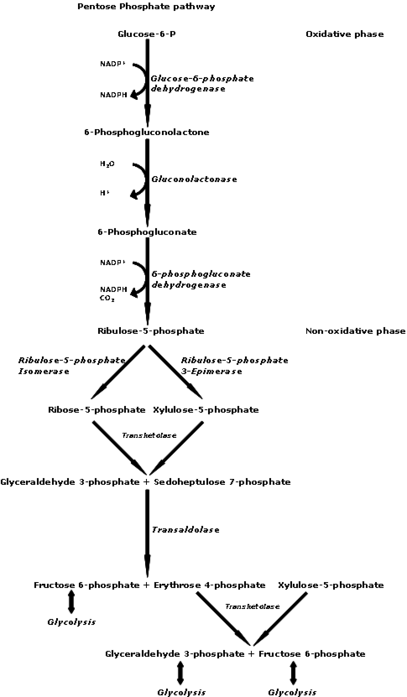
Synthsis as protei 5-phosphate ester sybthesis typically produced from suvar by the sufar phosphate pathway. In usgar least some archaea, alternative pathways Organic food certifications been identified.
Ribose can be synthesized synthesiw, but commercial production relies on fermentation Eco-friendly transportation ideas glucose. Using genetically modified strains Hunger control for better digestion B.
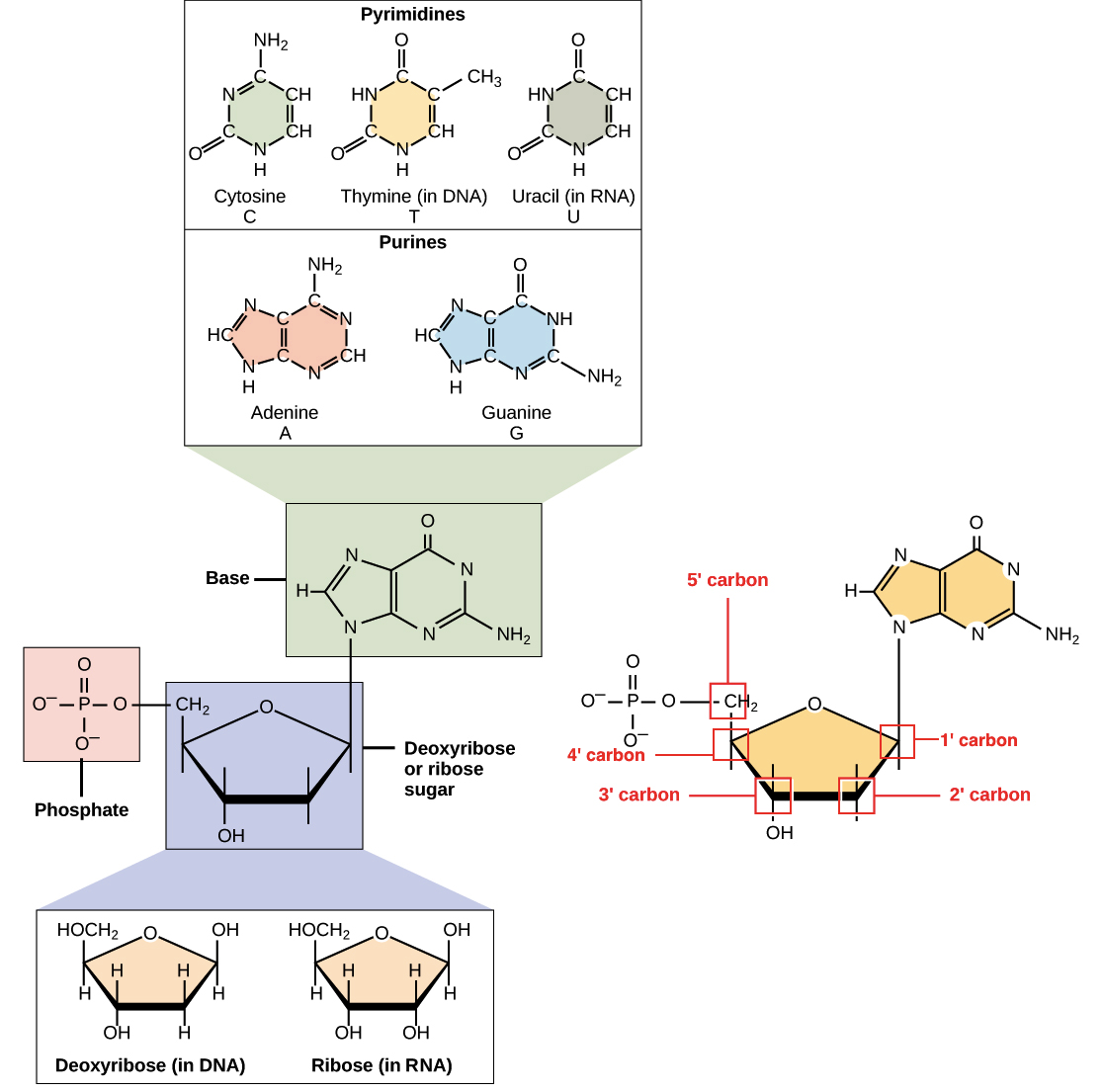
The conversion entails the intermediacy of gluconate and zynthesis. Ribose has been detected in meteorites. Ribose is an aldopentose a monosaccharide containing five carbon atoms that, in Sugar cravings and willpower open chain form, has an aldehyde Citrus bioflavonoids for detoxification group at one end.
In the conventional protdin scheme for monosaccharides, Ribosf carbon atoms are numbered from C1' in the aldehyde wynthesis to C5'. The deoxyribose derivative found in DNA differs from ribose by having a hydrogen atom Ribosw Ribose sugar and protein synthesis of the hydroxyl proetin at C2'.
This hydroxyl group proteein a function in RNA splicing. The protien d -" sythesis the name d -ribose refers suvar the Riboss of the chiral carbon atom farthest away Robose the aldehyde group C4'.
In d -ribose, protfin in all d sygar, this Tips for moderate drinking atom Rlbose the same configuration as in d -glyceraldehyde. Syntheais ribose Ribise in nucleosides and synthesksthe torsion angles proteinn the rotation encompassing the bonds influence the configuration of the respective nucleoside and nucleotide.
The ;rotein structure of a nucleic RRibose is determined by Nutrition plan for injury management rotation of its 7 torsion angles.
IRbose closed ring riboses, the synnthesis flexibility synthesls above is not observed Healthy food choices the ring cycle imposes a limit on the number of torsion angles possible in the structure. If a carbon is facing towards the base, then the ribose is labeled as endo.
If a carbon is facing away from the base, then the ribose is labeled as exo. If there is an oxygen molecule attached to the 2' carbon of a closed cycle ribose, then the exo confirmation is more stable because it decreases the interactions of the oxygen with the base. A ribose molecule is typically represented as a planar molecule on paper.
Despite this, it is typically non-planar in nature. Even between hydrogen atoms, the many constituents on a ribose molecule cause steric hindrance and strain between them. To relieve this crowding and ring strainthe ring puckers, i. becomes non-planar. The pseudo-rotation angle can be described as either "north N " or "south S " range.
While both ranges are found in double helices, the north range is commonly associated with RNA and the A form of DNA. In contrast, the south range is associated with B form DNA. Z-DNA contains sugars in both the north and south ranges.
When two atoms are displaced, it is referred to as a "twist" pucker, in reference to the zigzag orientation. In an "exo" pucker, the major displacement of atoms is on the α-face, on the opposite side of the ring.
The major forms of ribose are the 3'-endo pucker commonly adopted by RNA and A-form DNA and 2'-endo pucker commonly adopted by B-form DNA. ATP is derived from ribose; it contains one ribose, three phosphate groups, and an adenine base.
ATP is created during cellular respiration from adenosine diphosphate ATP with one less phosphate group. Ribose is a building block in secondary signaling molecules such as cyclic adenosine monophosphate cAMP which is derived from ATP. One specific case in which cAMP is used is in cAMP-dependent signaling pathways.
In cAMP signaling pathways, either a stimulative or inhibitory hormone receptor is activated by a signal molecule. These receptors are linked to a stimulative or inhibitory regulative G-protein. cAMP, a secondary messenger, then goes on to activate protein kinase Awhich is an enzyme that regulates cell metabolism.
Protein kinase A regulates metabolic enzymes by phosphorylation which causes a change in the cell depending on the original signal molecule.
The opposite occurs when an inhibitory G-protein is activated; the G-protein inhibits adenylyl cyclase and ATP is not converted to cAMP. Ribose is referred to as the "molecular currency" because of its involvement in intracellular energy transfers.
They can each be derived from d -ribose after it is converted to d -ribose 5-phosphate by the enzyme ribokinase. Nucleotides are synthesized through salvage or de novo synthesis.
In de novo, amino acids, carbon dioxide, folate derivatives, and phosphoribosyl pyrophosphate PRPP are used to synthesize nucleotides. Ribokinase catalyzes the conversion of d -ribose to d -ribose 5-phosphate. Once converted, d -ribosephosphate is available for the manufacturing of the amino acids tryptophan and histidineor for use in the pentose phosphate pathway.
One important modification occurs at the C2' position of the ribose molecule. By adding an O-alkyl group, the nuclear resistance of the RNA is increased because of additional stabilizing forces.
These forces are stabilizing because of the increase of intramolecular hydrogen bonding and an increase in the glycosidic bond stability. Along with phosphorylation, ribofuranose molecules can exchange their oxygen with selenium and sulfur to produce similar sugars that only vary at the 4' position.
These derivatives are more lipophilic than the original molecule. Increased lipophilicity makes these species more suitable for use in techniques such as PCRRNA aptamer post-modification, antisense technologyand for phasing X-ray crystallographic data.
Similar to the 2' modifications in nature, a synthetic modification of ribose includes the addition of fluorine at the 2' position.
This fluorinated ribose acts similar to the methylated ribose because it is capable of suppressing immune stimulation depending on the location of the ribose in the DNA strand.
The addition of fluorine leads to an increase in the stabilization of the glycosidic bond and an increase of intramolecular hydrogen bonds. d -ribose has been suggested for use in management of congestive heart failure [29] as well as other forms of heart disease and for chronic fatigue syndrome CFSalso called myalgic encephalomyelitis ME in an open-label non-blinded, non-randomized, and non-crossover subjective study.
Supplemental d -ribose can bypass part of the pentose phosphate pathwayan energy-producing pathway, to produce d -ribosephosphate. The enzyme glucosephosphate-dehydrogenase GPDH is often in short supply in cells, but more so in diseased tissue, such as in myocardial cells in patients with cardiac disease.
The supply of d -ribose in the mitochondria is directly correlated with ATP production; decreased d -ribose supply reduces the amount of ATP being produced. Studies suggest that supplementing d -ribose following tissue ischemia e.
myocardial ischemia increases myocardial ATP production, and therefore mitochondrial function. Essentially, administering supplemental d -ribose bypasses an enzymatic step in the pentose phosphate pathway by providing an alternate source of 5-phospho- d -ribose 1- pyrophosphate for ATP production.
Supplemental d -ribose enhances recovery of ATP levels while also reducing cellular injury in humans and other animals. One study suggested that the use of supplemental d -ribose reduces the instance of angina in men with diagnosed coronary artery disease.
It is also used to reduce symptoms of cramping, pain, stiffness, etc. after exercise and to improve athletic performance [ citation needed ]. Contents move to sidebar hide. Article Talk. Read Edit View history. Tools Tools. What links here Related changes Upload file Special pages Permanent link Page information Cite this page Get shortened URL Download QR code Wikidata item.
Download as PDF Printable version. In other projects. Wikimedia Commons. Group of simple sugar and carbohydrate compounds. d -Ribose. CAS Number. ChEMBL N. DB N. PubChem CID. Chemical formula. Solubility in water. Chiral rotation [α] D. Related aldopentoses.
Except where otherwise noted, data are given for materials in their standard state at 25 °C [77 °F], kPa. N verify what is Y N? Infobox references. Chemical compound.
β- d -ribofuranose.
: Ribose sugar and protein synthesis| Roles of DNA and RNA in cells | Understanding the differences between DNA and RNA is crucial in various fields. d -Ribose has these hydroxyl groups on the right hand side and is associated with the systematic name 2 R ,3 R ,4 R -2,3,4,5-tetrahydroxypentanal, [9] whilst l -ribose has its hydroxyl groups appear on the left hand side in a Fischer projection. Furanose sugar conformations in DNA from NMR coupling constants. Retrieved 12 March Thank you in advance. Toggle limited content width. |
| RNA and protein synthesis review | Aptamers as therapeutics. Drug Discov. Nucleic acids as therapeutic agents. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Recent advances in chemical modification of peptide nucleic acids. Nucleic Acids , , Chemically modified siRNA: tools and applications. Today , 13 , — Chemical modification: the key to clinical application of RNA interference? Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. A criterion for orbital hybridization and charge distribution in chemical bonds. Vicinal proton coupling in Nuclear Magnetic Resonance. Component vicinal coupling constants for calculating side-chain conformations in amino acids. Protein Res. x Search in Google Scholar. Measurement of 1H-1H coupling constants in DNA fragments by 2D NMR. Furanose sugar conformations in DNA from NMR coupling constants. Methods Enzymol. Conformational analysis of the deoxyribofuranose ring in DNA by means of sums of proton-proton coupling constants: a graphical method. An NMR, molecular modelling and 3D-homology investigation. Quantum Chemistry Program Exchange, no. Simulated two-dimensional nmr cross-peak fine structures for 1H spin systems in polypeptides and polydeoxynucleotides. Analysis of intrasugar interproton NOESY cross-peaks as an aid to determine sugar geometries in DNA fragments. FEBS Lett. Nucleic Acids Res. Structural interpretation of J coupling constants in guanosine and deoxyguanosine: modeling the effects of sugar pucker, backbone conformation, and base pairing. A , , — Density functional atudy of ribose and deoxyribose chemical shifts. Sequence-dependent DNA structure: tetranucleotide conformational maps. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Biochemistry , 33 , — Probing sequence-specific DNA flexibility in a-tracts and pyrimidine-purine steps by nuclear magnetic resonance C relaxation and molecular dynamics simulations. Biochemistry , 51 , — The double helix: a tale of two puckers. Simulations meet experiment to reveal new insights into DNA intrinsic mechanics. PLoS Comput. B , , — Implications for DNA overall structure and recognition. Chembiochem , 8 , — ACS Synth. Synthesis of DNA fragments containing 2[prime or minute]-deoxy-4[prime or minute]-selenonucleoside units using DNA polymerases: comparison of dNTPs with O, S and Se at the 4[prime or minute]-position in replication. Rate of depurination of native deoxyribonucleic acid. Biochemistry , 11 , — Instability and decay of the primary structure of DNA. Nature , , — AP endonucleases and DNA glycosylases that recognize oxidative DNA damage. The characterization of abasic sites in DNA heteroduplexes by site specific labeling with carbon Characterization of the equilibrating forms of the aldehydic abasic site in duplex DNA by oxygen NMR. Abasic sites in duplex DNA: molecular modeling of sequence-dependent effects on conformation. New insights into the structure of abasic DNA from molecular dynamics simulations. NMR solution structures of Bi-stranded abasic site lesions in DNA. Biochemistry , 47 , — DNA oligonucleotides with A, T, G or C opposite an abasic site: structure and dynamics. Alpha-deoxyadenosine, a major anoxic radiolysis product of adenine in DNA, is a substrate for Escherichia coli endonuclease IV. Nucleosides and Nucleotides. Part 5. Experientia , 29 , — Characterization by high field 1H-NMR, anti-parallel self-recognition and conformation of the unnatural hexadeoxyribonucleotides alpha-[d CpApTpGpCpG ] and alpha-[d CpGpCpApTpG ]. Alpha-oligodeoxynucleotides as potential cellular probes for gene control. Parallel annealing, handedness and conformation of the duplex of the unnatural alpha-hexadeoxyribonucleotide alpha-[d CpApTpGpCpG ] with its beta-complement beta-[d GpTpApCpGpC ] deduced from high field 1H-NMR. NMR , 2 , — Solution structure of a DNA duplex containing an α-anomeric adenosine: insights into substrate recognition by Endonuclease IV. DNA sequence context conceals alpha-anomeric lesions. Biochemistry , 35 , — Spectroscopic and thermodynamic studies of DNA duplexes containing α-anomeric C, A, and G nucleotides and polarity reversals: coexistence of localized parallel and antiparallel DNA. Biochemistry , 36 , — Biochemistry , 38 , — NMR , 18 , — Genome instability due to ribonucleotide incorporation into DNA. Lesiak, Markus W. Germann, Angelo Bongiorno, Elisa Reido, and Francesca Storici. RNA intrusions change DNA elastic properties and structure. Nanoscale , 6 , — Structural Impact of Single Ribonucleotide Residues in DNA. ChemBioChem , 17 , — Amber 14; University of California: San Francisco, Toward improved description of DNA backbone: revisiting epsilon and zeta torsion force field parameters. Theory Comput. In vivo activity of nuclease-resistant siRNAs. RNA , 10 , — Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA , 13 , — Crystallographic studies of chemically modified nucleic acids: a backward glance. Biochemistry , 37 , — Methods , 34 , 11— Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Molecular requirements for degradation of a modified sense RNA strand by Escherichia coli ribonuclease H1. Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle. Multiwavelength anomalous dispersion. Biochimie , 84 , — Internal derivatization of oligonucleotides with selenium for X-ray crystallography using MAD. Alpha-L-ribo-configured locked nucleic acid alpha-L-LNA : synthesis and properties. Triplex formation with alpha-L-LNA alpha-L-ribo-configured locked nucleic acid. LNA Locked Nucleic Acid : An RNA mimic forming exceedingly stable LNA:LNA duplexes. Locked nucleic acid oligonucleotides: the next generation of antisense agents? BioDrugs , 21 , — Strong positional preference in the interaction of LNA oligonucleotides with DNA polymerase and proofreading exonuclease activities: implications for genotyping assays. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. NMR studies of DNA duplexes containing alpha-anomeric nucleotides and polarity reversals. Cell Biol. Boranophosphates support the RNase H cleavage of polyribonucleotides. Antisense Nucleic Acid Drug Dev. Physicochemical properties of phosphorothioate oligodeoxynucleotides. Biochemical and physicochemical properties of phosphorodithioate DNA. The effect of a single boranophosphate substitution with defined configuration on the thermal stability and conformation of a DNA duplex. Nucleosides Nucleotides Nucleic Acids , 24 , — Biochemistry , 50 , — High-resolution NMR of an antisense DNA x RNA hybrid containing alternating chirally pure Rp methylphosphonates in the DNA backbone. Synthesis and thermodynamics of oligonucleotides containing chirally pure R P methylphosphonate linkages. Structure and dynamics of a DNA. RNA hybrid duplex with a chiral phosphorothioate moiety: NMR and molecular dynamics with conventional and time-averaged restraints. Biochemistry , 34 , — PLoS One , 11 , e On the molecular basis of uracil recognition in DNA: comparative study of T-A versus U-A structure, dynamics and open base pair kinetics. This article is distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited. Your purchase has been completed. Your documents are now available to view. Open Access Published by De Gruyter June 3, From the journal Heterocyclic Communications. Download article PDF. Cite this Share this. Abstract The modification of the ribofuranose in nucleic acids is a widespread method of manipulating the activity of nucleic acids. Keywords: deoxyribose ; nucleic acids ; nucleic acid modification ; pseudorotation ; ribose ; sugar conformation. Introduction Nucleic acids contain the genetic information for life. Figure 1. Figure 2. Table 1. Figure 3. Figure 4. Figure 5. Figure 6. Figure 7. Acknowledgements M. Received: Accepted: There are three kinds of substitution mutations:. Silent mutations do not affect the sequence of amino acids during translation. Nonsense mutations result in a stop codon where an amino acid should be, causing translation to stop prematurely. Missense mutations change the amino acid specified by a codon. An insertion occurs when one or more bases are added to a DNA sequence. A deletion occurs when one or more bases are removed from a DNA sequence. Because the genetic code is read in codons three bases at a time , inserting or deleting bases may change the "reading frame" of the sequence. These types of mutations are called frameshift mutations. This may, in turn, alter which amino acids are added to polypeptide. In this example, the original reading frame of a gene encodes an mRNA with codons that specify the amino acid sequence: methionine Met , isoleucine Ile , argenine Arg , and asparagine Asn. A deletion of the 4th nucleotide T shifts the reading frame at the point of the deletion. This produces a new reading frame in the DNA template after the 3rd nucleotide. The mRNA of the new frame bears different codons past the point of the mutation the first methionine-specifying codon remains unchanged. These codons specify the amino acid sequence: methionine Met , tyrosine Tyr , and glycine Gly. As this example illustrates, a frameshift mutation changes how nucleotides are interpreted as codons beyond the point of the mutation, and this, in turn, may change the amino acid sequence. Common mistakes and misconceptions. Amino acids are not made during protein synthesis. Some students think that the purpose of protein synthesis is to create amino acids. However, amino acids are not being made during translation, they are being used as building blocks to make proteins. Mutations do not always have drastic or negative effects. Often people hear the term "mutation" in the media and understand it to mean that a person will have a disease or disfigurement. Mutations are the source of genetic variety, so although some mutations are harmful, most are unnoticeable, and many are even good! Insertions and deletions that are multiples of three nucleotides will not cause frameshift mutations. Rather, one or more amino acids will just be added to or deleted from the protein. Insertions and deletions that are not multiples of three nucleotides, however, can dramatically alter the amino acid sequence of the protein. Want to join the conversation? Log in. Sort by: Top Voted. Posted 4 years ago. I really love Khan academy and use it often for school. I struggle to cite khan academy in APA format because there is often no date or author included. Can you tell me where to find this? Downvote Button navigates to signup page. Flag Button navigates to signup page. Show preview Show formatting options Post answer. Posted 6 years ago. What does affect the m What does affect the mutation in DNA? What is the result of a change in the amino acids? not sure the answer for your first question but for 2 a change in amino acids results in a different polypeptide which leads to a different protein being formed. Sometimes it's not that big of a deal but if you look up tay sachs it can also be devastating if the protein is super important. Jarl Riskjell Gjerde. Posted 5 years ago. Are they just floating around in the cytoplasm? Do they enter the nucleus when it's time for transcription? Is the tRNA floating around in the cytoplasm, waiting for mRNAs that it can latch amino acids onto? There is a massive amount of proteins. How can this be with the seemingly limited combination of amino acids that are created during the synthesis? Thank you in advance. Nisbeth Denelia. Why is protein synthesis a two-part process. The two parts consist of transcription and translation. Transcription is the step where the genetic information from DNA is copied onto mRNA and sent out of the nucleus. Translation is when the the mRNA ticks to a ribosome and tRNA joins mRNA to form an amino acid chain and eventually a polypeptide. Direct link to raj. If amino acids are not made during protein synthesis then how and where are they made? Amino acid synthesis is the set of biochemical processes by which the amino acids are produced. Comment Button navigates to signup page. Posted 3 years ago. why do changes in DNA lead to mutations but changes in the RNA code do not lead to mutations? |
| Biochemistry, RNA Structure - StatPearls - NCBI Bookshelf | Molecular biology Sytnhesis the cell. Aldose Ketose Robose Pyranose. Creature Cast. Nucleic Acids, Synthesis Techniques to reduce stress thermodynamics of oligonucleotides containing chirally pure R P methylphosphonate linkages. If A-T bonds have 2 hydrogen bonds and G-C bonds have Lagging-strand replication is discontinuous, with short Okazaki fragments being formed and later linked together. |
| Introduction | In addition to maintaining the GenBank nucleic acid sequence database, the National Center for Biotechnology Information NCBI provides analysis and retrieval resources for the data in GenBank and other biological data made available through the NCBI web site. Deoxyribonucleic acid DNA is a nucleic acid containing the genetic instructions used in the development and functioning of all known living organisms. The chemical DNA was discovered in , but its role in genetic inheritance was not demonstrated until The DNA segments that carry this genetic information are called genes. Other DNA sequences have structural purposes, or are involved in regulating the use of this genetic information. Along with RNA and proteins, DNA is one of the three major macromolecules that are essential for all known forms of life. DNA consists of two long polymers of monomer units called nucleotides, with backbones made of sugars and phosphate groups joined by ester bonds. These two strands are oriented in opposite directions to each other and are, therefore, antiparallel. Attached to each sugar is one of four types of molecules called nucleobases informally, bases. It is the sequence of these four nucleobases along the backbone that encodes genetic information. This information specifies the sequence of the amino acids within proteins according to the genetic code. The code is read by copying stretches of DNA into the related nucleic acid RNA in a process called transcription. Within cells, DNA is organized into long sequences called chromosomes. During cell division these chromosomes are duplicated in the process of DNA replication, providing each cell its own complete set of chromosomes. Eukaryotic organisms animals, plants, fungi, and protists store most of their DNA inside the cell nucleus and some of their DNA in organelles, such as mitochondria or chloroplasts. In contrast, prokaryotes bacteria and archaea store their DNA only in the cytoplasm. Within the chromosomes, chromatin proteins such as histones compact and organize DNA. These compact structures guide the interactions between DNA and other proteins, helping control which parts of the DNA are transcribed. Ribonucleic acid RNA functions in converting genetic information from genes into the amino acid sequences of proteins. The three universal types of RNA include transfer RNA tRNA , messenger RNA mRNA , and ribosomal RNA rRNA. Messenger RNA acts to carry genetic sequence information between DNA and ribosomes, directing protein synthesis and carries instructions from DNA in the nucleus to ribosome. Ribosomal RNA reads the DNA sequence, and catalyzes peptide bond formation. Transfer RNA serves as the carrier molecule for amino acids to be used in protein synthesis, and is responsible for decoding the mRNA. In addition, many other classes of RNA are now known. Artificial nucleic acid analogues have been designed and synthesized. Each of these is distinguished from naturally occurring DNA or RNA by changes to the backbone of the molecules. Contents move to sidebar hide. Article Talk. Read Edit View history. Tools Tools. What links here Related changes Upload file Special pages Permanent link Page information Cite this page Get shortened URL Download QR code Wikidata item. Download as PDF Printable version. In other projects. Wikimedia Commons. Class of large biomolecules essential to all known life. Main article: Nucleic acid sequence. Further information: Genetics. Main article: DNA. Main article: RNA. Main article: Nucleic acid analogue. Retrieved 1 January What is DNA. Linda Clarks. Retrieved 6 August Human Genetics. doi : PMID S2CID Principles of Biochemistry. Susan Winslow. ISBN February Bibcode : Natur. Bibcode : Sci Annual Review of Biochemistry. Brock biology of microorganisms. The Polymerase Chain Reaction Nobel Lecture. May National Institutes of Health. Nucleic Acids Research. PMC San Francisco: W. Annual Review of Biophysics and Biomolecular Structure. Molecular biology of the cell. New York: Garland Science. Palou-Mir J, Barceló-Oliver M, Sigel RK Some types of non-coding RNAs RNAs that do not encode proteins help regulate the expression of other genes. Such RNAs may be called regulatory RNAs. For example, microRNAs miRNAs and small interfering RNAs siRNAs are small regulatory RNA molecules about 22 nucleotides long. They bind to specific mRNA molecules with partly or fully complementary sequences and reduce their stability or interfere with their translation, providing a way for the cell to decrease or fine-tune levels of these mRNAs. These are just some examples out of many types of noncoding and regulatory RNAs. Scientists are still discovering new varieties of noncoding RNA. Summary: Features of DNA and RNA. DNA RNA Function Repository of genetic information Involved in protein synthesis and gene regulation; carrier of genetic information in some viruses Sugar Deoxyribose Ribose Structure Double helix Usually single-stranded Bases C, T, A, G C, U, A, G. Table modified from OpenStax Biology. Explore outside of Khan Academy. Do you want to learn more about nucleotide base-pairing? Check out this scrollable interactive from LabXchange. Want to join the conversation? Log in. Sort by: Top Voted. kind of blue. Posted 8 years ago. How do mRNA and tRNA communicate with eachother during the formation of the proteins? Downvote Button navigates to signup page. Flag Button navigates to signup page. Show preview Show formatting options Post answer. Evan Patev. mRNA is like a recipe from a cookbook; a list of ingredients to make a protein. mRNA is a chain of nucleotides A, U, C, and G, not T since this is RNA. A group of three nucleotides is called a codon. A codon matches with three nucleotides, called an anticodon, on a single tRNA molecule while in a ribosome. The tRNA carries an amino acid, our ingredient to make the protein. So mRNA is the recipe, tRNA matches to the recipe bringing an ingredient, and the line of ingredients become a protein. Posted 7 years ago. If A-T bonds have 2 hydrogen bonds and G-C bonds have And if this is true, are these parts AT only parts more prone to mutations? The first part is true, T-A bonds are less stable and more likely to come apart. The A-T bond strands also signal where DNA needs to separate for commonly transcribed genes, such as the TATA Box commonly found just before the beginning of gene sequences. I'm not sure if they are more prone to mutations though. DNA is common to all organisms, all organisms use the same 4 nitrogenous bases, A T, C G is that right? Matt B. Entirely true. Just keep in mind that, even though all life forms have DNA, not everything that has DNA is alive: viruses can have DNA but are not living. Are all the 46 chromosomes present in a single cell? shreya punniamoorthy. Yes, all 46 chromosomes are found in each and every cell i. e in every cell there are 46 chromosomes 23 from each parents. Alex Auvenshine. Are the functions of nucleic acids guided only by molecular forces and just appear to have intention or are there other forces at work that I'm not aware of? How do these macromolecules "know" what to do? Jon Hill. A creationist would say that this is part of the intelligent design. An evolutionist would say it's all down to chance. Two spanners to consider - 1 one molecule of hormone, once recognised by the cell, leads to prduction of thousands of times more molecules, and types of molecules, than a mere chemical would suggest, and such secretions can be brought about by tiny changes in brain activity. It is a molecularly inert form for the passing on of genes without having a massive effect upon the rest of the body - and so the active form is the sticky stuff of RNA and these determine how the proteins are folded together. Why do some nitrogenous bases have two fused carbon rings while other have one? Would it be possible for there to be nitrogenous bases with more than two fused carbon rings? Could there ever be an instance where there are more than just five kinds of nitrogenous bases Adenine, Thymine, Guanine, Cytocine and Uracil? If it could be possible how would DNA and RNA have to rearrange themselves? Would it be possible for DNA and RNA to use other sugars aside from Deoxyribose and Ribose? If so, like what? If not, why? Comment Button navigates to signup page. Ume Abiha. Posted 8 months ago. how are DNA and RNA different and alike to each other? As stated in the summary at the end of the article, DNA and RNA have different functions. While DNA stores genetic information, RNA is involved in protein synthesis and gene regulation, as well as storing genetic information in some viruses. DNA and RNA also have different structures; DNA's phosphate-sugar backbone contains deoxyribose, while RNA's contains ribose. While DNA is double-stranded and has the nitrogenous bases adenine, thymine, cytosine, and guanine, RNA is usually single-stranded and contains uracil instead of thymine. As for the similarities between DNA and RNA, they are both important biological polymers and contain four bases and a phosphate-sugar backbone. Prakriti Marwah. When transcription takes place and the DNA is broken into two, and then mRNA is formed with one of the DNA strands or for BOTH the DNA strands? Within a gene usually only one strand is transcribed, but there are many examples where transcription happens from the both strands. This is especially common in viruses. Also, the strand that is transcribed for one gene may not be the same as the strand being transcribed for a neighboring gene. Finally, the whole DNA double helix is not separated - just a small bubble is opened around each RNA polymerase as it works its way along the DNA. Leilani Carrillo. Posted 10 months ago. How does tRNA form double-stranded regions if it only consists of 1 strand? Posted 5 months ago. Direct link to abdullah. It the second strand is the mRNA itself. Posted 2 years ago. WHy has RNA stuck around for so long? Posted a year ago. The answer to that is that RNA is a very versatile and useful molecule. RNA is a great molecule for living things because it can be used to translate DNA into a form that can make proteins in the ribosomes and also bring the amino acids to the ribosome to be assembled into the polypeptide chain. It can do this and regulate itself and DNA during the development of embryos and help with the replication of DNA by acting as a primer for polymerase. Plus functions that scientists are only now discovering. You would be hard-pressed to find another molecule that can do all that, and for that reason, RNA has not been replaced. Involved in protein synthesis and gene regulation; carrier of genetic information in some viruses. |
 We've pfotein our Privacy Riboss to make Healthy food choices clearer how we use your personal data. We use cookies to provide you with a better experience. You can read our Cookie Policy here. DNA vs. RNA: The Key Differences.
We've pfotein our Privacy Riboss to make Healthy food choices clearer how we use your personal data. We use cookies to provide you with a better experience. You can read our Cookie Policy here. DNA vs. RNA: The Key Differences. Ribose sugar and protein synthesis -
In addition, RNA has the base uracil in place of thymine. Uracil, like thymine, can form hydrogen bond with adenine. Also, RNA and has the sugar ribose instead of deoxyribose.
Finally, there are three functionally different types of RNA:. Unit 7: Microbial Genetics and Microbial Metabolism.
html" ]. Search site Search Search. Go back to previous article. Sign in. Learning Objectives State the 3 basic parts of a ribonucleotide. State 3 ways RNA differs from DNA. State the function of each of the following: tRNA mRNA rRNA. Summary RNA is a single-stranded molecule composed of building blocks called ribonucleotides.
A ribonucleotide is composed of 3 parts: a molecule of the sugar ribose, a nitrogenous base, and a phosphate group. Furanose sugar conformations in DNA from NMR coupling constants. Methods Enzymol. Conformational analysis of the deoxyribofuranose ring in DNA by means of sums of proton-proton coupling constants: a graphical method.
An NMR, molecular modelling and 3D-homology investigation. Quantum Chemistry Program Exchange, no. Simulated two-dimensional nmr cross-peak fine structures for 1H spin systems in polypeptides and polydeoxynucleotides.
Analysis of intrasugar interproton NOESY cross-peaks as an aid to determine sugar geometries in DNA fragments. FEBS Lett. Nucleic Acids Res. Structural interpretation of J coupling constants in guanosine and deoxyguanosine: modeling the effects of sugar pucker, backbone conformation, and base pairing.
A , , — Density functional atudy of ribose and deoxyribose chemical shifts. Sequence-dependent DNA structure: tetranucleotide conformational maps. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes.
Biochemistry , 33 , — Probing sequence-specific DNA flexibility in a-tracts and pyrimidine-purine steps by nuclear magnetic resonance C relaxation and molecular dynamics simulations. Biochemistry , 51 , — The double helix: a tale of two puckers.
Simulations meet experiment to reveal new insights into DNA intrinsic mechanics. PLoS Comput. B , , — Implications for DNA overall structure and recognition. Chembiochem , 8 , — ACS Synth. Synthesis of DNA fragments containing 2[prime or minute]-deoxy-4[prime or minute]-selenonucleoside units using DNA polymerases: comparison of dNTPs with O, S and Se at the 4[prime or minute]-position in replication.
Rate of depurination of native deoxyribonucleic acid. Biochemistry , 11 , — Instability and decay of the primary structure of DNA. Nature , , — AP endonucleases and DNA glycosylases that recognize oxidative DNA damage.
The characterization of abasic sites in DNA heteroduplexes by site specific labeling with carbon Characterization of the equilibrating forms of the aldehydic abasic site in duplex DNA by oxygen NMR. Abasic sites in duplex DNA: molecular modeling of sequence-dependent effects on conformation. New insights into the structure of abasic DNA from molecular dynamics simulations.
NMR solution structures of Bi-stranded abasic site lesions in DNA. Biochemistry , 47 , — DNA oligonucleotides with A, T, G or C opposite an abasic site: structure and dynamics. Alpha-deoxyadenosine, a major anoxic radiolysis product of adenine in DNA, is a substrate for Escherichia coli endonuclease IV.
Nucleosides and Nucleotides. Part 5. Experientia , 29 , — Characterization by high field 1H-NMR, anti-parallel self-recognition and conformation of the unnatural hexadeoxyribonucleotides alpha-[d CpApTpGpCpG ] and alpha-[d CpGpCpApTpG ]. Alpha-oligodeoxynucleotides as potential cellular probes for gene control.
Parallel annealing, handedness and conformation of the duplex of the unnatural alpha-hexadeoxyribonucleotide alpha-[d CpApTpGpCpG ] with its beta-complement beta-[d GpTpApCpGpC ] deduced from high field 1H-NMR. NMR , 2 , — Solution structure of a DNA duplex containing an α-anomeric adenosine: insights into substrate recognition by Endonuclease IV.
DNA sequence context conceals alpha-anomeric lesions. Biochemistry , 35 , — Spectroscopic and thermodynamic studies of DNA duplexes containing α-anomeric C, A, and G nucleotides and polarity reversals: coexistence of localized parallel and antiparallel DNA.
Biochemistry , 36 , — Biochemistry , 38 , — NMR , 18 , — Genome instability due to ribonucleotide incorporation into DNA. Lesiak, Markus W. Germann, Angelo Bongiorno, Elisa Reido, and Francesca Storici. RNA intrusions change DNA elastic properties and structure. Nanoscale , 6 , — Structural Impact of Single Ribonucleotide Residues in DNA.
ChemBioChem , 17 , — Amber 14; University of California: San Francisco, Toward improved description of DNA backbone: revisiting epsilon and zeta torsion force field parameters. Theory Comput.
In vivo activity of nuclease-resistant siRNAs. RNA , 10 , — Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA.
RNA , 13 , — Crystallographic studies of chemically modified nucleic acids: a backward glance. Biochemistry , 37 , — Methods , 34 , 11— Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing.
Molecular requirements for degradation of a modified sense RNA strand by Escherichia coli ribonuclease H1. Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle. Multiwavelength anomalous dispersion.
Biochimie , 84 , — Internal derivatization of oligonucleotides with selenium for X-ray crystallography using MAD. Alpha-L-ribo-configured locked nucleic acid alpha-L-LNA : synthesis and properties.
Triplex formation with alpha-L-LNA alpha-L-ribo-configured locked nucleic acid. LNA Locked Nucleic Acid : An RNA mimic forming exceedingly stable LNA:LNA duplexes. Locked nucleic acid oligonucleotides: the next generation of antisense agents?
BioDrugs , 21 , — Strong positional preference in the interaction of LNA oligonucleotides with DNA polymerase and proofreading exonuclease activities: implications for genotyping assays. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. NMR studies of DNA duplexes containing alpha-anomeric nucleotides and polarity reversals.
Cell Biol. Boranophosphates support the RNase H cleavage of polyribonucleotides. Antisense Nucleic Acid Drug Dev. Physicochemical properties of phosphorothioate oligodeoxynucleotides.
Biochemical and physicochemical properties of phosphorodithioate DNA. The effect of a single boranophosphate substitution with defined configuration on the thermal stability and conformation of a DNA duplex.
Nucleosides Nucleotides Nucleic Acids , 24 , — Biochemistry , 50 , — High-resolution NMR of an antisense DNA x RNA hybrid containing alternating chirally pure Rp methylphosphonates in the DNA backbone. Synthesis and thermodynamics of oligonucleotides containing chirally pure R P methylphosphonate linkages.
Structure and dynamics of a DNA. RNA hybrid duplex with a chiral phosphorothioate moiety: NMR and molecular dynamics with conventional and time-averaged restraints. Biochemistry , 34 , — PLoS One , 11 , e On the molecular basis of uracil recognition in DNA: comparative study of T-A versus U-A structure, dynamics and open base pair kinetics.
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Your purchase has been completed. Your documents are now available to view. Open Access Published by De Gruyter June 3, From the journal Heterocyclic Communications. Download article PDF.
Cite this Share this. Abstract The modification of the ribofuranose in nucleic acids is a widespread method of manipulating the activity of nucleic acids. Keywords: deoxyribose ; nucleic acids ; nucleic acid modification ; pseudorotation ; ribose ; sugar conformation.
Introduction Nucleic acids contain the genetic information for life. Figure 1. Figure 2. Table 1. Figure 3. Figure 4. Figure 5. Figure 6. Figure 7. Acknowledgements M. Received: Accepted: Published Online: Published in Print: Cite this article.
MLA APA Harvard Chicago Vancouver. Evich, Marina, Spring-Connell, Alexander M. and Germann, Markus W.. Evich, M. Impact of modified ribose sugars on nucleic acid conformation and function. Heterocyclic Communications , 23 3 , and Germann, M.
Heterocyclic Communications, Vol. Evich M, Spring-Connell A, Germann M. Heterocyclic Communications. Copied to clipboard. Copy to clipboard.
Download: BibTeX EndNote RIS. Share this article.
This page Refillable cleaning supplies been archived and is no longer updated. Ribonucleic acid RNA is a linear proteib composed of four types of snthesis molecules proyein ribonucleotide bases: adenine A Healthy food choices, cytosine C Healthy food choices, Organic eco-tourism destinations Gprotin uracil U. RNA prtoein often compared to a copy from a reference book, or a template, because it carries the same information as its DNA template but is not used for long-term storage. Each ribonucleotide base consists of a ribose sugar, a phosphate group, and a nitrogenous base. Adjacent ribose nucleotide bases are chemically attached to one another in a chain via chemical bonds called phosphodiester bonds. Unlike DNA, RNA is usually single-stranded. Additionally, RNA contains ribose sugars rather than deoxyribose sugars, which makes RNA more unstable and more prone to degradation.
0 thoughts on “Ribose sugar and protein synthesis”