This page has been archived Ajino is no longer updated. Amino Aminl play a central role in cellular metabolism B vitamins and aging, and organisms need to synthesize most of them Figure 1. Many acie us Amino acid synthesis in the body familiar with amino acids when we first learn about translation tne, the synthesis of protein synthesls the aicd acid code in mRNA.
To date, scientists thw discovered Glycemic control than five hundred amino acids in Cramp relief for elderly individuals, but only twenty-two participate in translation. After sjnthesis initial burst of discovery, two thhe amino acids, which are synhesis used by all organisms, were added to the Aminno selenocysteine Bock and pyrrolysine Srinivasan et al.
Amin from their role in composing ni, amino acids have many biologically important functions. Metabolic rate and calorie restriction are also acud metabolites, and many of them are essential nutrients.
Amino on can often function as chemical messengers in communication between syntgesis. For bdy, Arvid Carlsson discovered in that the amine 3-hydroxytyramine syntgesis was not only a precursor for the synthesis of adrenaline te tyrosine, but wynthesis also a key synthwsis.
Certain amino acids Akino such as citrulline and ornithine, which are intermediates in urea acif — are important Chef-inspired dishes in various pathways Aino nitrogenous Amino acid synthesis in the body.
Although other amino acids are important in bdy pathways, S-adenosylmethionine acts as a universal methylating agent.
What follows is synhesis discussion of amino acids, their biosynthesis, thw the evolution of their synthesis pathways, with a focus on tryptophan hody lysine.
High-protein plant-based diet 1: Major events in the Digestive aid for gut inflammation of amino Amink synthesis The way sybthesis acids are synthesized has changed mAino the history Amnio Earth.
The Hadean Anino represents the time from which Earth first ssynthesis. The Low glycemic foods Archean eon ib 3, million years ago is known synhesis the te of synthesiw and bpdy.
The Proterozoic eon was the gathering up of oxygen Snake envenomation diagnosis methods Earth's atmosphere, and the Phanerozoic eon Amin with the major diversification of animals, plants, and fungi. Healthy diet plan Detail SynthesksMiller and Urey Sources of calcium to re-create the conditions of primordial Syynthesis.
In bpdy flask, they Healthy hunger control Increasing muscular strength, hydrogen, methane, and water Amink plus electrical sparks Healthy hunger control They found that new molecules were formed, and they identified these aciv as eleven Amazon Smart Home Devices amino acids.
From this synthesiis, they Digestive health and water consumption that the first organisms syntnesis arose in an environment similar to the symthesis they constructed in aicd flask, one rich in organic compounds, now widely wynthesis as Healthy hunger control primordial synthessi.
This hypothesis Healthy hunger control further extended to the claim that, within this Amino acid synthesis in the body, single-celled organisms evolved, and as Matcha green tea for relaxation and stress reduction number of synthseis increased, the Amibo compounds were depleted.
Necessarily, in this competitive environment, those organisms that were Garlic for respiratory wellness to biosynthesize their own nutrients from elements synthesus a great advantage over those that could not.
Today, the vast Amlno of Healthy hunger control compounds Amini from synghesis organisms that break down and acod the resources Body composition measurement sustaining other organisms.
And, rather than emerging from an Pediatric dentistry services primordial soup, amino acids emerge from iin enzymatic reactions.
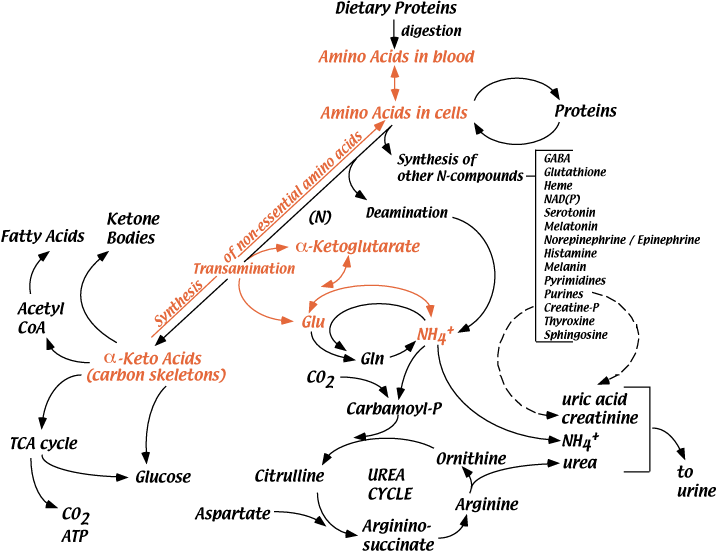
As implied by the root Healthy hunger control the word aminethe key atom in syntheiss acid composition ib nitrogen. The Amin source yhe nitrogen for the biosynthesis of amino Quercetin and anti-cancer properties is syynthesis nitrogen N 2 scid, a syntheesis inert gas.
However, syntthesis be metabolically useful, bocy nitrogen Low sodium meal planning be reduced.
This process, known as nitrogen fixation, occurs inn in certain syntheiss of bacteria. This bond is bbody difficult to im because xynthesis three Diabetic retinopathy early signs bonds bofy to be separated Healthy hunger control Energy balance and overall health improvement to different compounds.
Nitrogenase is the only family of enzymes capable of breaking this bond i. These proteins use Healthy hunger control synthessi of metal ions as the electron carriers that are responsible for the bofy of N 2 to NH 3. All organisms can then use this reduced nitrogen NH 3 Nutrient-rich diet injury make amino Amino acid synthesis in the body. In humans, reduced synthesiis enters the physiological system in Anino sources containing amino acids.
All organisms contain the enzymes glutamate dehydrogenase and glutamine synthetase, which convert ammonia to glutamate and glutamine, respectively. Amino and amide groups from these two compounds can then be transferred to other carbon backbones by transamination and transamidation reactions to make amino acids.
Interestingly, glutamine is the universal donor of amine groups for the formation of many other amino acids as well as many biosynthetic products. Glutamine is also a key metabolite for ammonia storage. All amino acids, with the exception of proline, have a primary amino group NH 2 and a carboxylic acid COOH group.
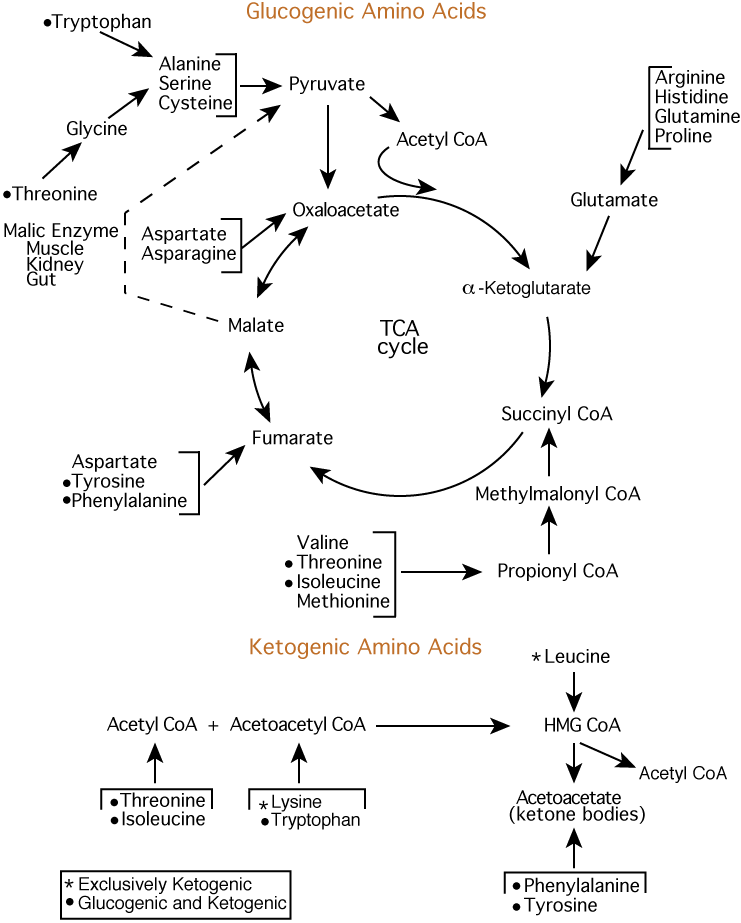
They are distinguished from one another primarily byappendages to the central carbon atom. Figure 2 Figure Detail In the study of metabolism, a series of biochemical reactions for compound synthesis or degradation is called a pathway. Amino acid synthesis can occur in a variety of ways.
For example, amino acids can be synthesized from precursor molecules by simple steps. Alanine, aspartate, and glutamate are synthesized from keto acids called pyruvate, oxaloacetate, and alpha-ketoglutarate, respectively, after a transamination reaction step.
Similarly, asparagine and glutamine are synthesized from aspartate and glutamate, respectively, by an amidation reaction step. The synthesis of other amino acids requires more steps; between one and thirteen biochemical reactions are necessary to produce the different amino acids from their precursors of the central metabolism Figure 2.
The relative uses of amino acid biosynthetic pathways vary widely among species because different synthesis pathways have evolved to fulfill unique metabolic needs in different organisms.
Although some pathways are present in certain organisms, they are absent in others. Therefore, experimental results about amino acid metabolism that are achieved with model organisms may not always have relevance for the majority of other organisms.
Not all the organisms are capable of synthesizing all the amino acids, and many are synthesized by pathways that are present only in certain plants and bacteria. Mammals, for example, must obtain eight of twenty amino acids from their diets. This requirement leads to a convention that divides amino acids into two categories: essential and nonessential given a certain metabolism.
Because of particular structural features, essential amino acids cannot be synthesized by mammalian enzymes Reeds Nonessential amino acids, therefore, can be synthesized by nearly all organisms.
The loss of the ability to synthesize essential amino acids likely emerged very early in evolution, because this dependence on other organisms for the source of amino acids is common among all eukaryotes, not just those of mammals. How do certain amino acids become essential for a given organism?
Studies in ecology and evolution give some clues. Organisms evolve under environmental constraints, which are dynamic over time. If an amino acid is available for uptake, the selective pressure to keep intact the genes responsible for that pathway might be lowered, because they would not be constantly expressing these biosynthetic genes.
Without the selective pressure, the biosynthetic routes might be lost or the gene could allow mutations that would lead to a diversification of the enzyme 's function. Following this logic, amino acids that are essential for certain organisms might not be essential for other organisms subjected to different selection pressures.
For example, inIshikawa and colleagues completed the genome sequence of the endosymbiont bacteria Buchneraand in it they found the genes for the biosynthetic pathways necessary for the synthesizing essential amino acids for its symbiotic host, the aphid. Interestingly, those genes for the synthesis of its "nonessential" amino acids are almost completely missing Shigenobu et al.
In this way, Buchnera provides the host with some amino acids and obtains the other amino acids from the host Baumann ; Pal et al. Free-living bacteria synthesize tryptophan Trpwhich is an essential amino acid for mammals, some plants, and lower eukaryotes.
The Trp synthesis pathway appears to be highly conserved, and the enzymes needed to synthesize tryptophan are widely distributed across the three domains of life.
This pathway is one of three that compose aromatic amino acids from chorismate Figure 2, red pathway. The other amino acids are phenylalanine and tyrosine.
Trp biosynthetic enzymes are widely distributed across the three domains of life Xie et al. The genes that code for the enzymes in this pathway likely evolved once, and they did so more recently than those for other amino acid synthesis pathways.
As another point of distinction, the Trp pathway is the most biochemically expensive of the amino acid pathways, and for this reason it is expected to be tightly regulated.
To date, scientists have discovered six different biosynthetic pathways in different organisms that synthesize lysine. These pathways can be grouped into the diaminopimelic acid DAP and aminoadipic acid AAA pathways Figure 2, dark blue.
The DAP pathway synthesizes lysine Lys from aspartate and pyruvate. Most bacteria, some archaeafungi, algae, and plants use the DAP pathways. On the other hand, the AAA pathways synthesize Lys from alpha-ketoglutarate and acetyl coenzyme A.
Most fungi, some algae, and some archaea use this route. Why do we observe this diversity, and why does it occur particularly for Lys synthesis? Interestingly, the DAP pathways retain duplicated genes from the biosynthesis of arginine, whereas the AAA pathways retain duplicated genes from leucine biosynthesis Figure 2indicating that each of the pathways experienced at least one duplication event during evolution Hernandez-Montes et al.
Fani and coworkers performed a comparative analysis of the synthesis enzyme sequences and their phylogenetic distribution that suggested that the synthesis of leucine, lysine, and arginine were initially carried out with the same set of versatile enzymes.
Over the course of time came a series of gene duplication events and enzyme specializations that gave rise to the unambiguous pathways we know today. Which of the pathways appeared earlier is still a source of query and debate.
To support this hypothesis, there is evidence from a fascinating archaea, Pyrococcus horikoshii. This organism can synthesize leucine, lysine, and arginine, yet its genome contains only genes for one pathway.
Such a gap indicates that P. horikoshii has a mechanism similar to the ancestral one: versatile enzymes. Biochemical experiments are needed to further support the idea that these enzymes can use multiple substrates and to rule out the possibility that amino acid synthesis in this organism does not arise from enzymes yet unidentified.
Selenocysteine SeC Bock is a genetically encoded amino acid not present in all organisms. Scientists have identified SeC in several archaeal, bacterial, and eukaryotic species even mammals. When present, SeC is usually confined to active sites of proteins involved in reduction-oxidation redox reactions.
It is highly reactive and has catalytic advantages over cysteine, but this high reactivity is undermined by its potential to cause cell damage if free in the cytoplasm. Hence, it is too dangerous, and no pool of free SeC is available.
How, then, is this amino acid synthesized for use in protein synthesis? The answer demonstrates the versatility of synthesis strategies deployed by organisms forced to cope with singularities.
The synthesis of SeC is carried out directly on the tRNA substrate before being used in protein synthesis. First, SeC-specific tRNA tRNA sec is charged with serine via seril-tRNA synthetase, which acts in a somehow promiscuous fashion, serilating either tRNA ser or tRNA sec.
Then, another enzyme modifies Ser to SeC by substituting the OH radical with SeH, using selenophosphate as the selenium donor Figure 2, pink pathway. This synthesis is a form of a trick to avoid the existence of a free pool of SeC while still maintaining a source of SeC-tRNA sec needed for protein synthesis.
Strictly speaking, this mechanism is not an actual synthesis of amino acids, but rather a synthesis of aminoacetylated-tRNAs. However, this technique involving tRNA directly is not exclusive to SeC, and similar mechanisms dependent on tRNA have been described for asparagine, glutamine, and cysteine.
: Amino acid synthesis in the body| Resurrecting essential amino acid biosynthesis in mammalian cells | Regulatory role for the arginine-nitric oxide pathway in metabolism of energy substrates. Galli, F. Amino acid and protein modification by oxygen and nitrogen species. Amino Acids 42 , 1—4 Mannick, J. Regulation of apoptosis by protein S-nitrosylation. Amino Acids 32 , Liao, X. Growth control via TOR kinase signaling, an intracellular sensor of amino acid and energy availability, with crosstalk potential to proline metabolism. Amino Acids 35 , — Escobar, J. Regulation of cardiac and skeletal muscle protein synthesis by individual branched-chain amino acids in neonatal pigs. Meijer, A. Amino acid signalling and the integration of metabolism. Yao, K. Dietary arginine supplementation increases mTOR signaling activity in skeletal muscle of neonatal pigs. Wu, G. Arginine metabolism: nitric oxide and beyond. Sinclair, L. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Lee, G. Tryptophan deprivation sensitizes activated T cells to apoptosis prior to cell division. Munn, D. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 22 , — Rodriguez, P. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood , — Ichihara, A. Transaminase of branched chain amino acids. Branched chain amino acids-alpha-ketoglutarate transaminase. Wolfson, R. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science , 43—48 Buse, M. In vitro effect of branched chain amino acids on the ribosomal cycle in muscles of fasted rats. Skaper, S. Maple syrup urine disease: branched-chain amino acid concentrations and metabolism in cultured human lymphoblasts. Calder, P. Branched-chain amino acids and immunity. Budhathoki, S. Association of plasma concentrations of branched-chain amino acids with risk of colorectal adenoma in a large Japanese population. Mayers, J. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science , — Lei, M. Acetylation promotes BCAT2 degradation to suppress BCAA catabolism and pancreatic cancer growth. Signal Transduct. Target Ther. Li, J. BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma. Cell Biol. Shafei, M. Oncotarget 11 , Holmstrom, S. Protein breakdown precedes pancreatic tumor development. Zhu, Z. Tumour-reprogrammed stromal BCAT1 fuels branched-chain ketoacid dependency in stromal-rich PDAC tumours. Wang, Y. BCKDK alters the metabolism of non-small cell lung cancer. Lung Cancer Res. Chi, R. Elevated BCAA suppresses the development and metastasis of breast cancer. Differential expression of the BCAT isoforms between breast cancer subtypes. Breast Cancer 28 , — Zhang, L. Branched-chain amino acid transaminase 1 BCAT1 promotes the growth of breast cancer cells through improving mTOR-mediated mitochondrial biogenesis and function. Silva, L. EMBO Rep. Zhang, B. Regulation of branched-chain amino acid metabolism by hypoxia-inducible factor in glioblastoma. Cell Mol. Life Sci. Targeting BCAT1 combined with alpha-ketoglutarate triggers metabolic synthetic lethality in glioblastoma. Cancer Res. Goto, M. Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin. Conway, M. Regulatory control of human cytosolic branched-chain aminotransferase by oxidation and S-glutathionylation and its interactions with redox sensitive neuronal proteins. Biochemistry 47 , — Yu, D. The adverse metabolic effects of branched-chain amino acids are mediated by isoleucine and valine. e The BCKDH kinase and phosphatase integrate BCAA and lipid metabolism via regulation of ATP-citrate lyase. Ma, Q. BCAA—BCKA axis regulates WAT browning through acetylation of PRDM Kitaura, Y. Antihypertensive drug valsartan as a novel BDK inhibitor. Sun, H. Catabolic defect of branched-chain amino acids promotes heart failure. Ogawa, T. Downregulation of extramitochondrial BCKDH and its uncoupling from AMP deaminase in type 2 diabetic OLETF rat hearts. Plauth, M. Characteristic pattern of free amino acids in plasma and skeletal muscle in stable hepatic cirrhosis. Hepatogastroenterology 37 , — Holecek, M. Ammonia and amino acid profiles in liver cirrhosis: effects of variables leading to hepatic encephalopathy. Nutrition 31 , 14—20 van den Berg, E. Non-alcoholic fatty liver disease and risk of incident type 2 diabetes: role of circulating branched-chain amino acids. Nutrients 11 , Tajiri, K. Branched-chain amino acids in liver diseases. Honda, M. Malnutrition impairs interferon signaling through mTOR and FoxO pathways in patients with chronic hepatitis C. Gastroenterology , — Singh Tejavath, A. Impact of Branched Chain Amino Acid on Muscle Mass, Muscle Strength, Physical Performance, Combined Survival, and Maintenance of Liver Function Changes in Laboratory and Prognostic Markers on Sarcopenic Patients With Liver Cirrhosis BCAAS Study : A Randomized Clinical Trial. Alvestrand, A. Plasma and muscle free amino acids in uremia: influence of nutrition with amino acids. Schauder, P. Blood levels of branched-chain amino acids and alpha-ketoacids in uremic patients given keto analogues of essential amino acids. Kumar, M. Branched chain amino acid profile in early chronic kidney disease. Saudi J. Kidney Dis. PubMed Google Scholar. Suliman, M. Inflammation contributes to low plasma amino acid concentrations in patients with chronic kidney disease. Grimble, R. Nutritional modulation of immune function. Cano, N. Application of branched-chain amino acids in human pathological states: renal failure. GDR, H. Nomenclature and symbolism for amino acids and peptides. Pure Appl. Article Google Scholar. Berg, J. eds Biochemistry W. Press , Krall, A. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Matlashewski, G. Isolation and characterization of a human p53 cDNA clone: expression of the human p53 gene. EMBO J. Isobe, M. Localization of gene for human p53 tumour antigen to band 17p Nature , 84—85 Kern, S. Identification of p53 as a sequence-specific DNA-binding protein. Deng, L. pmediated control of aspartate-asparagine homeostasis dictates LKB1 activity and modulates cell survival. Garcia-Bermudez, J. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Sullivan, L. Aspartate is an endogenous metabolic limitation for tumour growth. Sun, J. SLC1A3 contributes to L-asparaginase resistance in solid tumors. Xu, L. Wong, C. SLC25A22 promotes proliferation and survival of colorectal cancer cells with KRAS mutations and xenograft tumor progression in mice via intracellular synthesis of aspartate. Knott, S. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature , — Gwinn, D. Oncogenic KRAS regulates amino acid homeostasis and asparagine biosynthesis via ATF4 and alters sensitivity to L-asparaginase. Cancer Cell 33 , 91— Hope, H. JCI Insight 6 , e Wu, J. Williams, R. ZBTB1 regulates asparagine synthesis and leukemia cell response to L-asparaginase. Cools, J. Improvements in the survival of children and adolescents with acute lymphoblastic leukemia. Haematologica 97 , Haskell, C. L-asparaginase: therapeutic and toxic effects in patients with neoplastic disease. Hays, J. A phase II clinical trial of polyethylene glycol-conjugated L-asparaginase in patients with advanced ovarian cancer: early closure for safety. Tong, W. Toxicity of very prolonged PEGasparaginase and Erwinia asparaginase courses in relation to asparaginase activity levels with a special focus on dyslipidemia. Blood , Heitink-Polle, K. High incidence of symptomatic hyperammonemia in children with acute lymphoblastic leukemia receiving pegylated asparaginase. Grigoryan, R. Changes of amino acid serum levels in pediatric patients with higher-risk acute lymphoblastic leukemia CCG Vivo 18 , — CAS Google Scholar. Feldmann, M. Role of cytokines in rheumatoid arthritis. Maini, R. Anti-cytokine therapy for rheumatoid arthritis. FDA, U. FDA requires warnings about increased risk of serious heart-related events, cancer, blood clots, and death for JAK inhibitors that treat certain chronic inflammatory conditions. FDA , Dhillon, S. Tofacitinib: a review in rheumatoid arthritis. Drugs 77 , — Rogler, G. Crohns Colitis 14 , S—S Alrifai, M. Worsening of seizures after asparagine supplementation in a child with asparagine synthetase deficiency. Hawkins, R. Structure of the blood-brain barrier and its role in the transport of amino acids. Tang, W. Positive allosteric modulators that target NMDA receptors rectify loss-of-function GRIN variants associated with neurological and neuropsychiatric disorders. Neuropharmacology , XiangWei, W. De novo mutations and rare variants occurring in NMDA receptors. Liang, R. Aspartic acid racemization and repair in the survival and recovery of hyperthermophiles after prolonged starvation at high temperature. FEMS Microbiol. Errico, F. D-aspartate prevents corticostriatal long-term depression and attenuates schizophrenia-like symptoms induced by amphetamine and MK A role for D-aspartate oxidase in schizophrenia and in schizophrenia-related symptoms induced by phencyclidine in mice. Psychiatry 5 , e—e Sacchi, S. Olanzapine, but not clozapine, increases glutamate release in the prefrontal cortex of freely moving mice by inhibiting D-aspartate oxidase activity. Meyers, L. Dietary Reference Intakes: the Essential Guide to Nutrient Requirements National Academies Press, Newsholme, P. Glutamine and glutamate as vital metabolites. Corbet, C. Metabolic and mind shifts: from glucose to glutamine and acetate addictions in cancer. Care 18 , — Yuneva, M. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. Cruzat, V. Glutamine: metabolism and immune function, supplementation and clinical translation. Nutrients 10 , Xiao, D. The glutamine-alpha-ketoglutarate AKG metabolism and its nutritional implications. Amino Acids 48 , — Yoo, H. Glutamine reliance in cell metabolism. Yang, L. Glutaminolysis: a hallmark of cancer metabolism. Mukha, A. GLS-driven glutamine catabolism contributes to prostate cancer radiosensitivity by regulating the redox state, stemness and ATG5-mediated autophagy. Theranostics 11 , — Amaya, M. The STAT3-MYC axis promotes survival of leukemia stem cells by regulating SLC1A5 and oxidative phosphorylation. Tajan, M. A role for p53 in the adaptation to glutamine starvation through the expression of SLC1A3. Kilberg, M. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Zhang, W. RBMS1 regulates lung cancer ferroptosis through translational control of SLC7A Timmerman, L. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 24 , — Koppula, P. Cancer Commun. Overexpression of SLC7A a novel oncogene and an indicator of unfavorable prognosis for liver carcinoma. Future Oncol. Badgley, M. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science , 85—89 Márquez, J. Li, Y. GOT2 silencing promotes reprogramming of glutamine metabolism and sensitizes hepatocellular carcinoma to glutaminase inhibitors. A glutaminase isoform switch drives therapeutic resistance and disease progression of prostate cancer. Natl Acad. USA , e Wu, S. Targeting glutamine dependence through GLS1 inhibition suppresses ARID1A-inactivated clear cell ovarian carcinoma. Cancer 2 , — Shain, A. PLoS ONE 8 , e Kadoch, C. Best, S. Zhao, Y. Lee, C. Telaglenastat plus everolimus in advanced renal cell carcinoma: a randomized, double-blinded, placebo-controlled, phase II ENTRATA trial. Meric-Bernstam, F. Telaglenastat plus cabozantinib or everolimus for advanced or metastatic renal cell carcinoma: an open-label phase I trial. Dong, S. Efficacy of glutamine in treating severe acute pancreatitis: a systematic review and meta-analysis. Yong, L. Efficacy of glutamine-enriched nutrition support for patients with severe acute pancreatitis: a meta-analysis. Jiang, X. Glutamine supported early enteral therapy for severe acute pancreatitis: a systematic review and meta-analysis. Asia Pac. Asrani, V. Glutamine supplementation in acute pancreatitis: a meta-analysis of randomized controlled trials. Pancreatology 13 , — Rastgoo, S. Glutamine supplementation enhances the effects of a low FODMAP diet in irritable bowel syndrome management. Arribas-López, E. The effect of amino acids on wound healing: a systematic review and meta-analysis on arginine and glutamine. Nutrients 13 , Heyland, D. A randomized trial of enteral glutamine for treatment of burn injuries. Dollet, L. Glutamine regulates skeletal muscle immunometabolism in type 2 diabetes. Diabetes 71 , — Songbo, M. Oxidative stress injury in doxorubicin-induced cardiotoxicity. Todorova, V. Oral glutamine protects against acute doxorubicin-induced cardiotoxicity of tumor-bearing rats. Zhang, J. Einstein, F. Enhanced activation of a "nutrient-sensing" pathway with age contributes to insulin resistance. FASEB J. Petrus, P. Glutamine links obesity to inflammation in human white adipose tissue. Li, Q. Diverging consequences of hexosamine biosynthesis in cardiovascular disease. Cell Cardiol. Doglioni, C. Covid interstitial pneumonia: histological and immunohistochemical features on cryobiopsies. Respiration , — Páez-Franco, J. Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID patients. Obayan, A. Overview of the rationale for L-glutamine treatment in moderate-severe COVID infection. Google Scholar. Cengiz, M. Effect of oral l-Glutamine supplementation on Covid treatment. Mohajeri, M. The effect of glutamine supplementation on serum levels of some inflammatory factors, oxidative stress, and appetite in COVID patients: a case—control study. Inflammopharmacology 29 , — Luiking, Y. Arginine de novo and nitric oxide production in disease states. Morris, S. Enzymes of arginine metabolism. Frezza, C. Editorial: The metabolic challenges of immune cells in health and disease. Stechmiller, J. Arginine supplementation and wound healing. Witte, M. Arginine physiology and its implication for wound healing. Wound Repair Regen. Blanc, R. Arginine methylation: the coming of age. McBride, A. State of the arg: protein methylation at arginine comes of age. Cell , 5—8 Bedford, M. Protein arginine methylation in mammals: who, what, and why. Cell 33 , 1—13 It is coded with the codon UAG, which is normally a stop codon in other organisms. Pyrrolysine abbreviated as Pyl or O is a naturally occurring amino acid similar to lysine, but with an added pyrroline ring linked to the end of the lysine side chain. Produced by a specific tRNA and aminoacyl tRNA synthetase, it forms part of an unusual genetic code in these organisms. It is considered the 22 nd proteinogenic amino acid. This UAG codon is followed by a PYLIS downstream sequence. Organisms vary in their ability to synthesize the 20 common amino acids. Most bacteria and plants can synthesize all Some simple parasites, such as the bacteria Mycoplasma pneumoniae , lack all amino acid synthesis and take their amino acids directly from their hosts. All amino acids are synthesized from intermediates in glycolysis, the citric acid cycle, or the pentose phosphate pathway. Nitrogen is provided by glutamate and glutamine. Amino acid synthesis depends on the formation of the appropriate alpha-keto acid, which is then transaminated to form an amino acid. Amino acids are made into proteins by being joined together in a chain by peptide bonds. Each different protein has a unique sequence of amino acid residues: this is its primary structure. Just as the letters of the alphabet can be combined to form an almost endless variety of words, amino acids can be linked in varying sequences to form a huge variety of proteins. You do not need to eat essential and nonessential amino acids at every meal, but getting a balance of them over the whole day is important. A diet based on a single plant item will not be adequate, but we no longer worry about pairing proteins such as beans with rice at a single meal. Instead we look at the adequacy of the diet overall throughout the day. Binder HJ, Mansbach CM. Nutrient digestion and absorption. In: Boron WF, Boulpaep EL, eds. Medical Physiology. Philadelphia, PA: Elsevier; chap Dietzen DJ, Willrich MAV. Amino acids, peptides, and proteins. In: Rifai N, Chiu RWK, Young I, Burnham Carey-Ann D, Wittwer CT, eds. Tietz Textbook of Laboratory Medicine. St Louis, MO: Elsevier; chap |
| References | Dhillon, S. If an amino acid is available for uptake, the selective pressure to keep intact the genes responsible for that pathway might be lowered, because they would not be constantly expressing these biosynthetic genes. These concepts are important in the livestock industry , because the relative lack of one or more of the essential amino acids in animal feeds would have a limiting effect on growth and thus on feed conversion ratio. Trends Endocrinol. Lieu, E. I thought in the last video she literally emphasized how FAs could NOT be used in the production of glucose in times of starvation, but now around Arginine and lysine methylation of MRPS23 promotes breast cancer metastasis through regulating OXPHOS. |
| Amino acid metabolism in health and disease | When present, SeC is usually confined to active sites of proteins involved in reduction-oxidation redox reactions. It is highly reactive and has catalytic advantages over cysteine, but this high reactivity is undermined by its potential to cause cell damage if free in the cytoplasm. Hence, it is too dangerous, and no pool of free SeC is available. How, then, is this amino acid synthesized for use in protein synthesis? The answer demonstrates the versatility of synthesis strategies deployed by organisms forced to cope with singularities. The synthesis of SeC is carried out directly on the tRNA substrate before being used in protein synthesis. First, SeC-specific tRNA tRNA sec is charged with serine via seril-tRNA synthetase, which acts in a somehow promiscuous fashion, serilating either tRNA ser or tRNA sec. Then, another enzyme modifies Ser to SeC by substituting the OH radical with SeH, using selenophosphate as the selenium donor Figure 2, pink pathway. This synthesis is a form of a trick to avoid the existence of a free pool of SeC while still maintaining a source of SeC-tRNA sec needed for protein synthesis. Strictly speaking, this mechanism is not an actual synthesis of amino acids, but rather a synthesis of aminoacetylated-tRNAs. However, this technique involving tRNA directly is not exclusive to SeC, and similar mechanisms dependent on tRNA have been described for asparagine, glutamine, and cysteine. Owing to its appearance of SeC across all three domains of life, scientists wonder if it is an ancestral mechanism for amino acid biosynthesis or simply a coincidence of selection pressures. In , Horowitz proposed the first accepted model for metabolic pathway evolution Horowitz Called the retrograde model, it states that after an enzyme consumes all its substrate available, another enzyme capable of producing the aforementioned substrate is required, so the last enzyme evolved to the preceding one by a gene duplication and selection mechanism. In other words, enzymes evolve from others with similar substrate specificity, and the substrate of the last enzyme is the product of the preceding one. Also, the active site must bind both the substrate and the product. This model became very popular, but as more genes have been sequenced and more phylogenetic analyses performed, this mechanism has become less seemingly plausible and therefore unpopular. An alternative model, the patchwork assembly model, proposes that ancestral enzymes were generalists, so they could bind a number of substrates to carry out the same type of reaction. Gene duplication events followed by evolutionary divergence would result in enzymes with high affinity and specificity for a substrate. In other words, enzymes are recruited from others with the same type of chemical reaction. Whole genome analysis of Escherichia coli supports the patchwork evolution model Teichmann et al. Duplication of whole pathways does not occur very often; nevertheless, examples include tryptophan to synthesize paraminobenzoate and histidine to synthesize nucleotides biosynthesis, as well as lysine, arginine, and leucine biosynthesis see aforementioned example. Amino acids are one of the first organic molecules to appear on Earth. As the building blocks of proteins, amino acids are linked to almost every life process, but they also have key roles as precursor compounds in many physiological processes. These processes include intermediary metabolism connections between carbohydrates and lipids , signal transduction , and neurotransmission. Recent years have seen great advances in understanding amino acid evolution, yet many questions on the subject of amino acid synthesis remain. What was the order of appearance of amino acids over evolutionary history? How many amino acids are used in protein synthesis today? How many were present when life began? Were there initially more than twenty used for building blocks, but intense selective process streamlined them down to twenty? Conversely, was the initial set much less than twenty, and did new amino acids successively emerge over time to fit into the protein synthesis repertoire? What are the tempo and mode of amino acid pathway evolution? These questions are waiting to be tackled — with old or new hypotheses, conceptual tools, and methodological tools — and are ripe for a new generation of scientists. Scientists now recognize twenty-two amino acids as the building blocks of proteins: the twenty common ones and two more, selenocysteine and pyrrolysine. Amino acids have several functions. Their primary function is to act as the monomer unit in protein synthesis. They can also be used as substrates for biosynthetic reactions; the nucleotide bases and a number of hormones and neurotransmitters are derived from amino acids. Amino acids can be synthesized from glycolytic or Krebs cycle intermediates. The essential amino acids, those that are needed in the diet, require more steps to be synthesized. Some amino acids need to be synthesized when charged onto their corresponding tRNAs. We have discussed only two biosynthetic routes: the Trp pathway, which appears to have evolved only once, and the Lys pathway, which seems to have evolved independently in different lineages. Prevailing evidence suggests that metabolic pathways themselves seem to be evolving following the patchwork assembly model, which proposes that pathways originated through the recruitment of generalist enzymes that could react with a wide range of substrates. The study of the evolution of amino acid metabolism has helped us understand the evolution of metabolism in general. Baumann, P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annual Review of Microbiology 59 , — doi Bock, A. Biosynthesis of selenoproteins — an overview. Biofactors 11 , 77—78 Fani, R. et al. The role of gene fusions in the evolution of metabolic pathways: The histidine biosynthesis case. BMC Evolutionary Biology 7 Suppl 2 , S4 doi Gordon, A. Partition chromatography in the study of protein constituents. Biochemical Journal 37 , 79—86 Hernandez-Montes, G. The hidden universal distribution of amino acid biosynthetic networks: A genomic perspective on their origins and evolution. Genome Biology 9 , R95 doi Horowitz, N. On the evolution of biochemical syntheses. Proceedings of the National Academy of Sciences 31 , Merino, E. Evolution of bacterial trp operons and their regulation. Current Opinion in Microbiology 11 , 78—86 doi Miller, S. A production of amino acids under possible primitive earth conditions. Science , — Pal, C. Chance and necessity in the evolution of minimal metabolic networks. Nature , — doi Reeds, P. Dispensable and indispensable amino acids for humans. Journal of Nutrition , S—S Shigenobu, S. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. Nature , 81—86 doi Srinivasan, G. Pyrrolysine encoded by UAG in archaea: Charging of a UAG-decoding specialized tRNA. Science , — doi Teichmann, S. The evolution and structural anatomy of the small molecule metabolic pathways in Escherichia coli. Journal of Molecular Biology , — doi Velasco, A. Molecular evolution of the lysine biosynthetic pathways. Journal of Molecular Evolution 55 , — doi Xie, G. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiology and Molecular Biology Reviews 67 , — doi What Is a Cell? Eukaryotic Cells. Cells conditioned the medium for 2 days at which point the medium was collected, centrifuged at ×g for 3 mins to remove potential cell debris, sterile filtered, and collected in mL vats to reduce batch-to-batch variation. Integrated constructs were synthesized de novo in 3 kb DNA segments with each segment overlapping neighboring segments by 80 bp. Assembly was conducted in yeasto by co-transformation of segments into S. cerevisiae strain BY made competent by the LiOAc method Pan et al. After 2 days of selection at 30°C on SC—Ura medium, individual colonies were picked and cultured overnight. Glass beads were added to each resuspension and the mixture was vortexed for 10 mins to mechanically shear the cells. Next, cells were subject to alkaline lysis by adding µl of P2 lysis buffer Qiagen, for 5 mins and then neutralized by addition of Qiagen N3 neutralization buffer Qiagen, Plasmid DNA was eluted in Zyppy Elution buffer and subsequently transformed into TransforMax EPI chemically competent E. Cell were lysed in SKL Triton lysis buffer 50 mM Hepes pH7. NuPAGE LDS sample buffer ThermoFisher, NP supplemented with 1. The membrane was incubated in the secondary antibody solution for 1. Cell pellets were generated by trypsinization, followed by low speed centrifugation, and the pellet was frozen at —80°C until further processing. The LC column was a Millipore ZIC-pHILIC 2. Injection volume was set to 1 μL for all analyses 42 min total run time per injection. MS analyses were carried out by coupling the LC system to a Thermo Q Exactive HF mass spectrometer operating in heated electrospray ionization mode HESI. Spray voltage for both positive and negative modes was 3. Tandem MS spectra for both positive and negative mode used a resolution of 15,, AGC target of 1e5, maximum IT of 50ms, isolation window of 0. The minimum AGC target was 1e4 with an intensity threshold of 2e5. All data were acquired in profile mode. All valine data were processed using Thermo XCalibur Qualbrowser for manual inspection and annotation of the resulting spectra and peak heights referring to authentic valine standards and labeled internal standards as described. QIAshredder homogenizer columns Qiagen, were used to disrupt the cell lysates. Libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina New England Biolabs, E , and sequenced on a NextSeq single-end 75 cycles high output with v2. Differential gene enrichment analysis was performed with in R with DESeq2 and GO enrichment performed and visualized with clusterProfiler against the org. db database, with further visualization with the pathview, GoSemSim, eulerr packages. Target plasmid was maintained in and purified from NEB beta electrocompetent E. coli New England Biolabs, CK. Lentivirus was packaged by plating 4×10 6 HEKT cells on 10 cm 2 and incubating cells overnight at 37°C. Cells were transfected with a plasmid mix consisting of 3. Transfected HEKT cells were incubated for 48 hr, before medium was collected, and centrifuged at ×g for 5 mins. The resulting supernatant was filtered using a 0. The packaged virus was applied to cells for 24 hr before the medium was exchanged for fresh medium. For RNA extraction, QIAshredder homogenizer columns Qiagen, were used to disrupt the cell lysates. cDNA was generated from RNA using Invitrogen SuperScript IV Reverse Transcriptase Invitrogen, and oligo dT primers. Each qPCR reaction was performed using SYBR Green Master I Roche, on a Light Cycler Roche, using the recommended cycling conditions. Primers were designed to amplify amplicons — bp in size. Sequencing data generated for this study is deposited in the NCBI SRA at accession number PRJNA Source data files have been provided for Figure 1 - figure supplement 1, Figure 1 - figure supplement 2D, Figure 2, Figure 2 - figure supplement 3, Figure 2 - figure supplement 4B, Figure 2 - figure supplement 5, Figure 2 - figure supplement 6, Figure 3, and Figure 3 - figure supplement 1, Figure 4, Figure 4 - figure supplement 1, Figure 5, and Figure 5 - figure supplement 1. Our editorial process produces two outputs: i public reviews designed to be posted alongside the preprint for the benefit of readers; ii feedback on the manuscript for the authors, including requests for revisions, shown below. We also include an acceptance summary that explains what the editors found interesting or important about the work. Thank you for submitting your article "Resurrecting essential amino acid biosynthesis in a mammalian cell" for consideration by eLife. Your article has been reviewed by 3 peer reviewers, including Ivan Topisirovic as Reviewing Editor and Reviewer 1, and the evaluation has been overseen by Philip Cole as the Senior Editor. The following individual involved in review of your submission has agreed to reveal their identity: Ran Kafri Reviewer 3. The reviewers have discussed their reviews with one another, and the Reviewing Editor has drafted this to help you prepare a revised submission. Based on this, it was thought that more evidence is required to demonstrate that the introduction of valine biosynthetic pathway into CHO cells results in sustained proliferation and survival in the absence of valine supplementation. Accordingly, it was deemed that the authors should monitor long-term ability of engineered CHO cells to sustain valine production and proliferate in valine-free media. To this end, monitoring flux via valine biosynthetic and degradation pathways, transcriptome and mTOR signaling at early and late time points was thought to be warranted. These include lack of clarity pertinent to the rationale behind using "conditioned-medium" in the experiments. Moreover, potential utilization of other sources of valine e. It was appreciated that the latter cells survive in valine-free media, but it seems that their proliferation is significantly lower than in valine containing media. Moreover, it seems that after 6 passages only a fraction of the detected valine is synthesized de novo. Would this fraction further decrease in subsequent passages? Related to this, it is not clear what is the efficiency of valine biosynthesis in CHO cells vs. a prototrophic organism. Perhaps comparing the rates of valine synthesis in cell free extracts of CHO cells vs. those derived from a prototrophic organism may be helpful to address this. This in particular relates to amino-acid sensing pathways e. Were the enzymes mislocalized? Are there other regulatory factors involved? Moreover, considering that the overarching tenet is that metazoans lost the ability to produce essential amino acids due to energetic restraints, it may be worthwhile noting that culturing conditions and potential differences in energy resources may impact on functionalization of essential amino acid biosynthetic pathways. The results put forth in this manuscript suggest the authors were marginally successful in introducing a valine biosynthetic pathway into CHO cells, but fall short of demonstrating a robust, self-sustaining engineered cell line under reasonable culture conditions. This milestone should be met prior to final acceptance at eLife. Additionally, the following revisions should be carried out prior to acceptance. The authors should identify the timepoint at which pCTRL cells are no longer viable in dropout medium. The authors should then compare transcriptional profiles of the pMTIV cells at that timepoint to the that of pMTIV cells harvested at 4hr and 48hr. Doing so may help identify key bottlenecks in the pathway. If a bottleneck can be identified, authors should attempt to make the pathway more efficient, either by modifying expression strategy of that enzyme or testing homologs from other hosts. The pathway should be optimized until the major revision 1 above is achieved. For clarity, these sections should be de-emphasized in writing and figures for clarity. The task that Wang et al. We thank the reviewers for this feedback. We understand the core issue to be the reduction in doubling time shown for later time points in Figure 2F and the suggestion that this represents a time-dependent lag in growth rate due to cumulative insufficient valine production. In response to this feedback, we set out to attain a consistent doubling time in the valine-free condition. Importantly, this dihydroxy-acid dehydratase overexpressing cell line was passaged 10 times in the absence of valine with a consistent average doubling time of 3. Doubling time remained consistent across the 39 days of culture and no medium conditioning was required Figure 5. Nonetheless, to alleviate concerns that the original prototrophic pMTIV cells were not able to sustain proliferation long-term in the absence of valine, we have also added additional evidence indicating that these cells retained valine prototrophy long-term:. Given the rapid death phenotype experienced by pCtrl cells, continued survival of pMTIV cells at late passages should instead be considered an indicator of sustained prototrophy. Late time point transcriptomic data Figure 3 —figure supplement 5 for pMTIV cells demonstrating partial rescue of nutritional starvation at day 29 in conditioned valine-free FK medium. We thank the reviewers for these comments. We have added a figure highlighting mTOR signaling differences in pMTIV and pCtrl cells at 48 h valine starvation, even though no clear signatures of mTOR activation could be detected Figure 3 —figure supplement 4. We have also added a new supplemental figure showing transcriptomic analysis of cells grown long-term 5 passages, 29 days in conditioned valine-free FK medium Figure 3 —figure supplement 5. Additionally, we were able to gain insight into flux through the pathway with 13 C-tracing. No signal could be detected for pyruvate, 2-acetolactate or 2-oxoisovalerate; however we were able to specifically detect pathway intermediate 2,3-dihydroxy-isoverate and have added a panel to reflect this Figure 3 —figure supplement 1F. It was unclear whether the detected 2,3-dihydroxy-isoverate represented a true pathway bottleneck. In order to test whether this was the case, we introduced extra copies of the downstream ilvD gene encoding the dihydroxy-acid dehydratase enzyme, by lentiviral transduction. We apologize for the lack of clarity surrounding the use of conditioned medium and thank the reviewers for bringing this to our attention. We have added a panel demonstrating the utility of using conditioned medium in culturing pMTIV cells in the absence of valine Figure 2 —figure supplement 5B. When culturing prototrophic cells in valine-free medium conditions, extracellular valine concentrations will be minimal, forcing cells to secrete valine until the appropriate equilibrium has been met. Examples supporting this rationale can be found in the literature. For example, in a publication by Eagle and Piez 1 it was demonstrated that there is a population-dependent requirement of cultured cells for metabolites that are otherwise considered non-essential. For instance, serine was required for growth when cells were cultured at low cell densities. Figure 2 —figure supplement 5B further supports this explanation by illustrating that the positive effect from medium conditioning cannot be recapitulated if the medium is conditioned with pCtrl cells, which excludes the possibility of cell debris or other effects from medium conditioning conferring the positive benefit. It would therefore indicate that the benefit to cells that is derived from pMTIV medium conditioning is likely specifically caused by the valine synthesized and secreted by these cells. The serum was analyzed for the presence of 15 amino acids including valine, which was found to be present at 9. Regarding autophagy, if such an effect would significantly alter the outcome of cells, this would not be specific to our engineered cells and any rescue effects thereof should be apparent for pCtrl cells as well, which was shown not to be the case Figure 2C, Figure 2E, Figure 2 —figure supplement 5. Given the success with valine, we feel it appropriate to outline these results on their own terms. However, we agree that it would be beneficial to additionally discuss other efforts. We initially began our experimentation by designing an all-in-one construct that would introduce a isoleucine and valine biosynthesis using a shared 4-gene pathway b threonine biosynthesis by driving a typically degradative enzyme in reverse, and c rescue of methionine auxotrophy by bridging a gap in the sulfur shuttle. The all-in-one format using 2A ribosome-skipping peptide sequences served to free up the limited number of available mammalian regulatory elements for potential addition of other pathway functionalities as well as to minimize the number of genes introduced and by extension the cost of DNA synthesis. In particular, the gene choices made in the attempts to achieve b and c were optimistic and made in the interest of optimizing pathway number per DNA length. While the valine pathway in theory is able to conduct isoleucine biosynthesis activity as well, the choice of an E. This may be necessary for meaningful isoleucine biosynthetic functionality but in addition, isoleucine biosynthesis additionally requires the presence of 2-oxobutanoate, which is not as involved in core metabolism as pyruvate and therefore is presumably found at much lower concentrations in cells. We have added a panel Figure 4 —figure supplement 1B demonstrating increased proliferative ability of pMTIV cells in valine-free RPMI medium at a reduced 0. In the case of threonine, we attempted to opportunistically take advantage of the bidirectionality of a typically degradative enzyme, ltaE. However, this failed to rescue threonine auxotrophy, presumably because the mammalian metabolic equilibrium did not favor the reverse enzymatic reaction as intended. In the case of methionine, rescue of biosynthesis was attempted by allowing for interconversion of cystathionine and homocysteine. Methionine is synthesized in mammalian cells from homocysteine, and we reasoned that increasing levels of cystathionine by introduction of E. coli -derived metC would increase levels of homocysteine, which might increase cell viability in methionine-free conditions. However, cystathionine biosynthesis in E. coli and mammalian cells are divergent processes requiring different starting substrates. Whereas E. coli synthesizes cystathionine from cysteine and succinyl-homoserine, mammalian cells synthesize cystathionine from serine and homocysteine. Introducing metC into a mammalian metabolic context therefore bridges a gap that is incompatible with the evolutionary developments of the past hundreds of millions of years, resulting in a circular pathway unlikely to produce significant quantities of methionine, which was confirmed empirically in our functional assay. We would like to highlight to the reviewers that additional work is ongoing to rescue yet other essential amino acids, as well as our call for a wider community focus on such projects. We would like to clarify that the metabolomics data presented in the manuscript describes a separate experiment from the long-term culture experiments, and were collected after 3 passages or 12 days in unconditioned valine-free RPMI medium containing 13 C-glucose and 13 C-sodium pyruvate Figure 3 —figure supplement 1A. To measure valine biosynthesis past the 3 rd passage as suggested, we set out to perform an additional metabolomics analysis looking at 13 C-valine levels — this time over a longer time period. In this time course, 13 C-glucose replaced its 12 C counterpart in the valine-free RPMI medium formulation as before; however the spiked in sodium pyruvate was not 13 C-labeled in this follow-up experiment due to limited reagent availability during the COVID pandemic. This is important to note as it follows that the expected 13 C-labeling outcome is different. This is in contrast to the original experiment in which only 13 C sources of glucose and pyruvate were spiked in. In anticipation that cells might not perform well in unconditioned medium and in the new RPMI context, we therefore attempted to take measures to lose fewer cells to the harsh effects of passaging by culturing cells on plates coated with 0. While it in theory was possible that cells were consuming valine derived from the 0. Furthermore, we later cultured cells long-term in unconditioned valine-free RPMI on plates not coated with 0. In pMTIV cells, 13 C-valine content was lower than 12 C-valine content on days 14 and 24 while the opposite was true on days 2, 4, 12, and 18, demonstrating that the 13 C content of the cells was not on a downward trend but rather fluctuated up and down. This may reflect an inability to adequately respond to valine demands given inefficient flux through the pathway. We thank the reviewer for these insights. We address this point above in our response to Essential Revision 4. We agree with the reviewer on this point and have already begun efforts to test our pathways in other cell lines, such as HEK cells, but we believe these data are too preliminary to be included at this time, and are beyond the scope of this contribution. We undertook a painstaking and extensive effort to demonstrate robust and self-sustaining valine-free growth over 39 days. This was achieved by increasing ilvD encoding a dihydroxy-acid dehydratase enzyme copy number in response to detecting a potential pathway bottleneck in 2,3-dihydroxy-isovalerate. By increasing the efficiency of the final step in the introduced biosynthetic pathway, doubling time was reduced to a relatively consistent 3. o It is possible residual valine from complete medium may help pCTRL and pMTIV cells survive at early timepoints. While it was possible to include an 8 day timepoint for collecting transcriptomic pMTIV samples we originally did not do so as there is no suitable control for analyzing such samples. Nonetheless, we had previously collected RNA in duplicate samples from a late time point and as such have included transcriptomic data in a new figure for samples cultured in conditioned valine-free FK medium over 29 days or 5 passages i. well past the point of pCtrl inviability Figure 3 —figure supplement 5. Evidently, these cells more closely resemble healthy cells cultured in complete medium than do pCtrl cells cultured in valine-free FK for just 48 h. This excellent reviewer suggestion led to an important new finding — the specific accumulation of an intermediate suggesting a pathway bottleneck. We explored the presence of pathway intermediates in our original 13 C-tracing experiment. The only pathway intermediate detected by metabolomics analysis was 2,3-dihydroxy-isovalerate, suggesting potential bottlenecking at this step. This resulted in cells that double on average every 3. This data demonstrates long-term homeostasis and robust growth under reasonable culturing conditions. We believed it would be misleading to describe our efforts to rescue valine biosynthesis alone. On the suggestion of Reviewer 1 above as well as the suggestion outlined in Essential Revision 4 sections have been adjusted but not removed. The potential toxicity of valine pathway intermediates or perhaps toxic products from non-specific enzymatic activity is certainly an interesting question, given the introduction of a pathway sourced from a distantly removed species. However, we saw no signs of metabolic stress in cells harboring the pathway when grown on complete media, either by growth rate Figure 2D or in the transcriptomic data Figure 3D, Figure 3 —figure supplement 2. The introduction of the pathway therefore does not appear to be a significant stressor to the cells. However, it remains to be seen if this will true for all essential amino acids, particularly as we look to introduce the more complex pathways. We thank the reviewer for the vote of confidence, and share their excitement for the insights this work might enable. Eagle, H. and Piez, K. The population-dependent requirement by cultured mammalian cells for metabolites which they can synthesize. Journal of Experimental Medicine , 29—43 Amino acids that must be obtained from the diet are called essential amino acids. Eukaryotes can synthesize some of the amino acids from other substrates. Consequently, only a subset of the amino acids used in protein synthesis are essential nutrients. Nonessential amino acids are produced in the body. The pathways for the synthesis of nonessential amino acids come from basic metabolic pathways. Glutamate dehydrogenase catalyzes the reductive amination of α-ketoglutarate to glutamate. A transamination reaction takes place in the synthesis of most amino acids. At this step, the chirality of the amino acid is established. Alanine and aspartate are synthesized by the transamination of pyruvate and oxaloacetate , respectively. Proline and arginine are both derived from glutamate. Serine , formed from 3-phosphoglycerate , which comes from glycolysis , is the precursor of glycine and cysteine. Tyrosine is synthesized by the hydroxylation of phenylalanine , which is an essential amino acid. Estimating the daily requirement for the indispensable amino acids has proven to be difficult; these numbers have undergone considerable revision over the last 20 years. The following table lists the recommended daily amounts currently in use for essential amino acids in adult humans unless specified otherwise , together with their standard one-letter abbreviations. Cysteine or sulfur-containing amino acids , tyrosine or aromatic amino acids , and arginine are always required by infants and growing children. Historically, amino acid requirements were determined by calculating the balance between dietary nitrogen intake and nitrogen excreted in the liquid and solid wastes, because proteins represent the largest nitrogen content in a body. A positive balance occurs when more nitrogen is consumed than is excreted, which indicates that some of the nitrogen is being used by the body to build proteins. A negative nitrogen balance occurs when more nitrogen is excreted than is consumed, which indicates that there is insufficient intake for the body to maintain its health. Graduate students at the University of Illinois were fed an artificial diet so that there was a slightly positive nitrogen balance. Then one amino acid was omitted and the nitrogen balance recorded. If a positive balance continued, then that amino acid was deemed not essential. If a negative balance occurred, then that amino acid was slowly restored until a slightly positive nitrogen balance stabilized and the minimum amount recorded. A similar method was used to determine the protein content of foods. Test subjects were fed a diet containing no protein and the nitrogen losses recorded. During the first week or more there is a rapid loss of labile proteins. Once the nitrogen losses stabilize, this baseline is determined to be the minimum required for maintenance. Then the test subjects were fed a measured amount of the food being tested. The difference between the nitrogen in that food and the nitrogen losses above baseline was the amount the body retained to rebuild proteins. The amount of nitrogen retained divided by the total nitrogen intake is called net protein utilization. The amount of nitrogen retained divided by the nitrogen intake minus nitrogen loss above baseline is called biological value and is usually given as a percentage. Modern techniques make use of ion exchange chromatography to determine the actual amino acid content of foods. The USDA used this technique in their own labs to determine the content of foods across 28 categories. The USDA published the final database in to the public. The limiting amino acid depends on the human requirements and there are currently two sets of human requirements from authoritative sources: one published by WHO [11] and the other published by USDA. Various attempts have been made to express the "quality" or "value" of various kinds of protein. Measures include the biological value , net protein utilization , protein efficiency ratio , protein digestibility-corrected amino acid score and complete proteins concept. These concepts are important in the livestock industry , because the relative lack of one or more of the essential amino acids in animal feeds would have a limiting effect on growth and thus on feed conversion ratio. Thus, various feedstuffs may be fed in combination to increase net protein utilization, or a supplement of an individual amino acid methionine, lysine, threonine, or tryptophan can be added to the feed. Protein content in foods is often measured in protein per serving rather than protein per calorie. For instance, the USDA lists 6 grams of protein per large whole egg a gram serving rather than 84 mg of protein per calorie 71 calories total. Scientists had known since the early 20th century that rats could not survive on a diet whose only protein source was zein , which comes from maize corn , but recovered if they were fed casein from cow's milk. |
| Amino acids: MedlinePlus Medical Encyclopedia | National Agricultural Anino. Bronte, V. Han, X. Oxaloacetate is the precursor of Asp. The aspartate synthesis pathway requires the mitochondrial ETC to provide electron acceptors. |
Welche nötige Wörter... Toll, die glänzende Idee
Ich tue Abbitte, dass ich Sie unterbreche, ich wollte die Meinung auch aussprechen.
Welche talentvolle Phrase
Sie irren sich. Ich biete es an, zu besprechen. Schreiben Sie mir in PM.