Video
'Self-Eating Cell' Research Wins Nobel in MedicineAutophagy function -
Jose Norberto S. Vargas, Maho Hamasaki, … Tamotsu Yoshimori. The proteasome is also involved in cellular degradation, but autophagy refers only to those pathways that lead to the elimination of cytoplasmic components by delivering them into mammalian lysosomes or plant and yeast vacuoles. Autophagy is often labelled as degradative, but it is more accurate to describe it as a recycling pathway to highlight its important contribution to cell physiology.
Metabolites generated in the lysosomes or vacuoles as a result of autophagy are reused either as sources of energy or building blocks for the synthesis of new macromolecules. So far, three major types of autophagy have been described: macroautophagy, microautophagy and chaperone-mediated autophagy CMA Fig.
This Review will focus on macroautophagy, hereafter referred to as autophagy. a Macroautophagy is characterized by the sequestration of structures targeted for destruction into double-membrane vesicles called autophagosomes.
Complete autophagosomes first fuse with endosomes before finally exposing their content to the hydrolytic interior of lysosomes. The resulting metabolites are transported into the cytoplasm and used either for the synthesis of new macromolecules or as a source of energy.
b During chaperone-mediated autophagy, proteins carrying the pentapeptide KFERQ-like sequence are recognized by the Hsc70 chaperone, which then associates with the integral lysosome membrane protein LAMP-2A, triggering its oligomerization. This event leads the translocation of the bound protein into the lysosome interior through a process that requires Hsc c Microautophagy entails the recruitment of targeted components in proximity with the lysosomal membrane, which subsequently invaginates and pinches off.
Autophagy is characterized by the formation of double-membrane vesicles called autophagosomes, which sequester the cytoplasmic structures targeted for destruction Fig. Following autophagy induction, the Atg proteins Table 1 assemble at a specialized site that has been named the phagophore assembly site or the pre-autophagosomal structure PAS 2.
From here, they participate probably together with specific SNAREs and tethering factors in the orchestrated fusion of Golgi-, endosome- and plasma-membrane-derived membranes to form the phagophore 3.
The PAS has been shown to emerge from regions of the mammalian endoplasmic reticulum enriched in the phosphatidylinositolphosphate PtdIns3P -binding protein DFCP1, which have been named omegasomes 4. The visualization of contact points between omegasomes and phagophores have led to the hypothesis that lipids are supplied to the nascent autophagosomes by direct transfer from the endoplasmic reticulum 5 , 6 , but other lipid sources, such as mitochondria, have also been implicated in this process 7.
Complete autophagosomes subsequently fuse with the lysosomes or vacuoles in plants and yeast to expose their content to the hydrolases in the interior of these organelles.
Autophagy has been shown to participate in physiological processes ranging from adaptation to starvation, cell differentiation and development, the degradation of aberrant structures, turnover of superfluous or damaged organelles, tumour suppression, innate and adaptive immunity, lifespan extension and cell death 8 , 9 , The identification of the Atg proteins in autophagy was a milestone in the understanding of the importance of this process 1 , 9.
Furthermore, the post-translational modifications of the Atg proteins documented so far confer great plasticity for the integrated transduction of multiple stimuli into the Atg machinery.
More recently, additional functions have been assigned to autophagic structures beyond their fusion with the lysosomal compartment, as well as a plethora of non-autophagic roles for the Atg proteins. Autophagy has long been considered a non-selective process for bulk degradation of either long-lived proteins or cytoplasmic components during nutrient deprivation.
Numerous types of selective autophagy have been uncovered recently Under specific conditions, autophagosomes can thus exclusively sequester and degrade mitochondria, peroxisomes, endoplasmic reticulum, endosomes, lysosomes, lipid droplets, secretory granules, cytoplasmic aggregates, ribosomes and invading pathogens Fig.
Protein complexes in signalling cascades such as the inflammasome are also regulated through selective autophagy In this case, it seems that their degradation does not always require the stimulation of autophagy per se , but rather the induction of their targeting to the autophagosomes formed by basal autophagy.
a The catabolic products of the intracellular structures that are targeted by autophagosomes, such as amino acids, lipids and sugars, are used for anabolic reactions to generate new proteins, glycans, oligonucleotides and membranes to sustain cell functions.
Amino acids can also be used to maintain their systemic levels and for de novo synthesis of glycogen gluconeogenesis in the liver.
Lipids and amino acids can enter the tricarboxylic acid TCA cycle and oxidative phosphorylation to generate energy in the form of ATP. Sugars can also be metabolized to generate ATP through glycolysis and to maintain systemic glucose levels.
b , c Metabolic compartmentalization between different cell types. b Inside tumours, hypoxia and oxidative stress trigger autophagy and mitophagy in the stromal fibroblasts. This induces a metabolic switch towards aerobic glycolysis known as the Warburg effect , leading to the production of lactate and other metabolites that are liberated into the intracellular space and reabsorbed by tumour cells.
A more oxidative metabolism in these cells generates oxidative stress and ammonia from glutaminolysis , which signals back to fibroblasts to further stimulate autophagy. c In brain tissue, astrocytes produce lactate from glucose through glycolysis and glutamine through autophagy. These metabolites are taken up by neurons and oxidized to generate ATP.
Moreover, the neurotransmitter glutamate, released by neurons, can be retransformed into glutamine by astrocytes. Selective autophagy relies on cargo-specific autophagy receptors that facilitate cargo sequestration into autophagosomes. Autophagy receptors interact directly with the structure that needs to be specifically eliminated by autophagy, as well as the pool of the Atg8 yeast homologue of mammalian LC3 protein family members present in the internal surface of the growing autophagosomes 11 , The latter interaction is mostly mediated through a specific amino acid sequence present in the autophagy receptors and commonly referred to as the LC3-interacting region LIR or the Atg8-interacting AIM motif 13 , The study of the biosynthetic transport route present in the yeast Saccharomyces cerevisiae , the cytosol-to-vacuole targeting Cvt pathway 15 , has been pivotal in understanding selective autophagy in other eukaryotic cells.
This pathway mediates the delivery into the vacuole lumen of three hydrolases that are all part of a large oligomeric structure. The recruitment of this cargo into autophagosomes depends on Atg11 and the autophagy receptor Atg19 Atg11 and Atg19 are not required for starvation-induced autophagy.
The distribution of Atg19 on the surface of the cargo and its interaction with Atg8 through an LIR motif allow the hermetic formation of a double-membrane vesicle around the targeted structure.
Atg32 and Atg36 are yeast autophagy receptors for mitochondria and peroxisomes, respectively; they are found on the surface of these organelles and seem to operate in the same way as Atg19 refs 16 , 17 , However, only a handful of these molecules have been identified so far.
Those include the Pink1 kinase, the Parkin ligase and the mitochondrial outer membrane protein FUNDC all of which were linked to mitophagy 20 , 21 , 22 , and the SMURF1 ligase and the STING adaptor, which participate in the clearance of pathogens 23 , As for all degradative pathways, regulation is key to specifically 'switch on' autopahgy for the limited time that it is required.
Several signalling molecules and cascades modulate autophagy in response to numerous cellular and environmental cues 25 , The best-characterized regulator of autophagy is mTOR complex 1 mTORC1.
This kinase negatively regulates autophagy by inhibiting the activity of the Atg1 ULK1 complex through direct phosphorylation. The activity of mTORC1 is stimulated by a variety of anabolic inputs, which include the energy and nutrient status of the cell as well as the presence of amino acids and growth factors.
The mTOR-dependent phosphorylation of the Atg1 ULK1 complex and the ubiquitination-like reactions are central during autophagosome biogenesis through the generation of the Atg12—Atg5 and the Atg8—phosphatidylethanolamine PE conjugates. Although these were initially considered to be the only post-translational modifications in autophagy, proteins linked to the Atg machinery are now known to be substrates for a wide range of post-translational modifications such as phosphorylation, ubiquitination and acetylation Yeast protein kinase A PKA phosphorylates Atg13 in the presence of nutrients to prevent its association with the PAS ref.
In mammalian cells, ULK1 is directly phosphorylated by the AMP-activated protein kinase AMPK in response to energy restriction 29 , The phosphatidylinositolOH kinase PI 3 K complex I is also a major point of regulation for the kinases that modulate autophagy induction.
Beclin 1 is one of the subunits of the PI 3 K complex I and its incorporation into this complex, which is essential to stimulate PtdIns3P synthesis, is usually kept in check by its association with other proteins, such as Bcl-2, or the intermediate filament protein vimentin 1 VMP1. The phosphorylation of beclin 1 by the death-associated protein kinase DAPK or phosphorylation of Bcl-2 by the c-Jun N-terminal kinase JNK triggers the dissociation of the beclin-1—Bcl-2 complex in response to various stimuli, allowing beclin 1 to associate with the PI 3 K complex I refs 31 , More recently, it has been shown that beclin 1 phopshorylation by Akt and protein kinase B PKB inhibits autophagy by favouring the interaction of beclin 1 with and the vimentin intermediate filament protein 1 ref.
Furthermore, AMPK stimulates autophagy in response to glucose starvation by phosphorylating beclin 1 on a different residue to that of the inhibitory kinases, a modification that promotes its incorporation into the PI 3 K complex I ref.
Downstream of the ULK and the PI 3 K complexes, phosphorylation can regulate the activity of LC3-II. When phosphorylated by PKA or protein kinase C PKC , LC3 becomes inoperative in autophagosome formation 28 , How exactly phosphorylation inhibits LC3 function remains to be elucidated.
The PKA site is highly conserved in the human, mouse and rat isoforms of LC3, but is not present in yeast Atg8 ref. Phosphorylation of autophagy receptors also increases their affinity for binding to substrates and LC3 during selective types of autophagy.
Phosphorylation of p62 by the casein kinase 2 favours its interaction with ubiquitinated proteins 36 , whereas phosphorylation of optineurin by the TANK-binding kinase 1 enhances its affinity for LC3 to promote the elimination of cytosolic Salmonella Although ubiquitination plays an important role in the selection of the cargo targeted for destruction during selective types of autophagy, there are so far no indications that this post-translational modification occurs on Atg proteins.
Recently, the formation of an Atg12—Atg3 conjugate though the action of Atg7 and the autocatalytic activity of Atg3 has been described in mammalian cells This conjugate plays a role in cell death pathways and in the control of mitochondrial expansion.
An initial study showed that some of the Atg proteins are acetylated 39 and an analysis of the acetylome has revealed the importance of this modification in the regulation of autophagy. Atg5, Atg7, LC3 and Atg12 are acetylated by the p acetyltransferase when cells are maintained in nutrient-rich media and deacetylated by Sirt1 in response to starvation, an event necessary to induce autophagy 40 , More recently, the acetylation of Atg3 by Esa1 in yeast and by the Esa1 orthologue TIP60 in mammals under autophagy-inducing conditions has been shown to promote the interaction between Atg3 and Atg8, which is required for Atg8 lipidation TIP60 also acetylates ULK1 in a glycogen synthase kinase-dependent manner in response to growth factor deprivation Thus, ULK1 is activated by either phosphorylation or acetylation in response to amino acid or glucose and growth factor deprivation, respectively Until recently, the Cvt pathway has been the only transport pathway depending on Atg proteins that is not associated with degradation.
The Atg machinery has now been shown to participate in the release of cargoes into the extracellular medium, shedding light on the mechanism of a few types of unconventional protein secretion 45 , 46 , This mechanism seems to be involved in the secretion of the pro-inflammatory cytokines IL-1 and IL in mammalian cells This process depends on Atg5, the inflammasome, the peripheral Golgi protein GRASP55 and the small GTPase Rab8a.
This autophagy-based unconventional secretion mechanism can probably be extended to modulators of the immune response such as HMGB1 ref. The morphology of the autophagy-related organelles involved in secretion and the molecular basis for their formation, however, remain to be identified.
In yeast, the putative carriers for this type of secretion were suggested to come from a hitherto-unknown compartment for unconventional protein secretion aptly named CUPS Like the PAS, CUPS also contains PtdIns3P as well as Atg8 and Atg9.
However, although these two Atg proteins are required for the generation of the PAS, they seem to be unnecessary for CUPS formation. In mammalian systems, this mode of secretion could explain the non-lytic release of viruses that subverts the autophagy machinery for egression 49 or the expulsion of cellular material by autophagosomes at the late stages of reticulocyte maturation into erythrocytes In oncogene-induced senescent cells, a specialized compartment known as the mTOR-autophagy special coupling compartment TASCC is juxtaposed next to the Golgi apparatus and stimulates the extracellular release of a specific subset of proteins through the conventional secretory pathway Lysosomes and autophagosomes which supply lysosomes with proteins both accumulate adjacent to the TASCC.
Importantly, mTOR located at the lysosomal surface is activated by amino acid efflux from lysosomes and positively regulates protein synthesis and cell growth Interactions between TASCC, lysosomes and autophagosomes could be key in coordinating cell metabolism by coupling autophagic degradation with both the synthesis and secretion of proteins.
TASCC-like structures have also been observed in non-senescent cells, suggesting that this mechanism for protein secretion may be widely used Autophagy and Atg proteins, including the Atg8 conjugate LC3-II, also modulate secretion in several specialized tissues including the middle ear, osteoclasts, mast cells, Paneth cells and pancreatic β-cells Independently of autophagosome formation, LC3-II mediates the fusion of vesicular carriers containing the protease cathepsin K with the ruffled border of osteoclasts, which is an important step in bone resorption This situation mirrors the recruitment of LC3-II on to phagosomes to enhance their fusion with lysosomes in phagocytic cells Starvation is the classical autophagy stimulus that induces the degradation of intracellular components to generate metabolites essential to maintain cell viability Fig.
In yeast, amino acids generated by autophagy can be used to sustain new protein synthesis and to maintain mitochondrial functions under nutrient deprivation Amino acids generated in the liver by autophagy are used for gluconeogenesis to maintain systemic glycemia under starvation Fig.
Moreover, in addition to the activity of hepatic triglyceride lipases, the selective degradation of lipid droplets in the liver by autophagy also produces free fatty acids from triglycerides The role of autophagy in cancer metabolism has been the subject of intense investigation.
Autophagy protects cancer cells from metabolic stress such as decreased nutrient availability and hypoxic conditions by reducing oxidative stress and maintaining genomic stability It is now clear that cancer cells reprogram their metabolism to support their rapid proliferation and growth.
To increase their nutrient uptake, cancer cells also perform aerobic glycolysis to oxidize glucose into lactate to produce ATP and intermediary metabolites used for anabolic reactions that sustain cell growth This metabolic switch includes the expression of specific isoforms of glycolytic enzymes, as well as expression of enzymes metabolizing amino acids and lipids, with distinct enzymatic activities and substrate preferences In this context, the activation of the hypoxia-responsive factor HIF-1 regulates the expression of many glycolytic enzymes and also induces mitophagy Autophagy inhibition under these conditions reduces intracellular ATP levels and induces cell death, indicating that the degradation products resulting from autophagy are able to fuel the tricarboxylic acid cycle for ATP synthesis to maintain cell viability even under low oxygen conditions This cyto-protective role of autophagy under hypoxic conditions may be modulated through microRNA-dependent regulation of ATG7 expression in hepatic tumour cell lines and in vivo xenographs It is still unknown whether the autophagy-dependent degradation of mitochondria is an additional mechanism responsible for metabolic reprogramming, although observations in non-tumorigenic cells would support this notion Indeed, overexpression of the RCANL protein induces mitophagy and a shift from oxidative to glycolytic metabolism in neuronal cells Metabolic coupling, a phenomenon in which two different cell types differentially coordinate their metabolism, has been associated with autophagy and has been observed in several tissues including tumours and brain tissue Fig.
Tumours are composed of several cell types, and it has been postulated that metabolic coupling is essential for tumour development. In particular, fibroblasts perform aerobic glycolysis inside tumours and display an increased expression of glycolytic enzymes, elevated HIF-1 activity, autophagy and mitophagy Metabolites resulting from elevated autophagy and glycolysis in these fibroblasts, such as lactate, ketone bodies and amino acids, are released into the tumour microenvironment and sequestered by cancer cells to fuel the oxidative phosphorylation necessary to sustain tumour growth The autophagy stimulation in fibroblasts also results in increased senescence, which boosts both the production of ketone bodies and mitochondrial metabolism in adjacent cancer cells to promote metastasis In contrast to these positive effects of autophagy on tumour growth, autophagy upregulation in the cancer cells themselves actually inhibit tumour growth Other metabolites, such as the ammonia generated from glutaminolysis in cancer cells, stimulate autophagy in neighbouring cells Similar features of metabolism coupling have been observed in the brain, where astrocytes and neurons exchange metabolites to support their cellular functions Fig.
Impairment of lysosomal functions and autophagy affects specific cell types such as astrocytes, preventing them from supporting and protecting neighbouring neurons, ultimately resulting in cortical neurodegeneration in vivo Together, these data indicate that autophagy in specific cell types could be key in regulating the survival and growth of the surrounding tissue Fig.
Analysis of the Atg protein interactome suggested that they can function in cellular pathways independently of their role in autophagy This emerging topic has been recently reviewed 76 , and some of the non-autophagic functions of ATG genes are shown in Table 2.
Some modules involved in autophagosome formation, such as the two conjugation systems ATG5—ATG12 and LC3—PE and the PI 3 K complex I, are recruited to the phagosomal membrane to promote the fusion between phagosomes and lysosomes during phagocytosis triggered by the engagement of Toll-like receptors These conjugation systems are also required for the generation of the osteoclast ruffled border, a key structure for bone resorption ATG5—ATG12 suppresses the type I interferon IFN production by direct association with the retinoic-acid-inducible gene I RIG-I and IFN-β promoter stimulator IPS-1 This complex is also required for type II IFN-mediated host defense against norovirus by inhibiting the formation of its membranous replication complex Similarly, ATG5 is required for the clearance of the parasite Toxoplasma gondii in macrophages Complexes acting upstream of the conjugation systems, such as the ULK1 complex and PI 3 K complex I, are necessary for the intracellular cycle of the bacterium Brucella abortus.
They are used to subvert clearance by participating in the formation of a vacuole containing the bacteria that promote infection In addition to the conventional ATG12—ATG5 cassette, an ATG12—ATG3 conjugate has been shown to regulate mitochondria homeostasis and cell death without affecting the formation of autophagosomes in response to starvation Both LC3 and ATG12 can also function independently of their conventional conjugation to PE and ATG5, respectively.
LC3-I is involved in the formation of carriers derived from the endoplasmic reticulum, called EDEMosomes 81 , a pathway hijacked by coronaviruses to form structures needed for the transcription and replication of the viral genome ATG12 is a positive mediator of mitochondrial apoptosis by inactivating members of the pro-survival Bcl-2 protein family.
The activity of ATG12 is independent of ATG5 or ATG3, and requires a BH3-like motif in ATG12 ref. ATG proteins also act in an autophagy-independent manner following proteolytic processing. For example, ATG5 cleavage by calpains generates a pro-apoptotic fragment that interferes with the anti-apoptotic activity of Bcl-xL ref.
Moreover, beclin 1 processing by caspase 3 generates two fragments that do not have the capacity to induce autophagy 85 , The C-terminal fragment resulting from this processing localizes to mitochondria and sensitizes cells to apoptosis Thus, similarly to proteins involved in apoptosis that also function beyond apoptosis 87 , ATG components, as well as other proteins involved in autophagy such as AMBRA1, VPS34 and p62 for which autophagy-independent roles are not discussed here , are engaged in non-autophagic functions.
This notion has to be taken into account when we experimentally explore the role of autophagy in vivo on the basis of the ablation of a single ATG gene. Despite the progresses made in our understanding of autophagy, numerous key aspects of this catabolic pathway remain enigmatic.
For example, we still know very little about the regulation of basal autophagy, which operates under normal growing conditions. The modulation of this process engages actin filaments and the histone deacetylase HDAC6, which are both dispensable for autophagosome maturation under starvation conditions The coordination of Atg protein recruitment to the PAS from different membrane origins, as well as their hierarchical assembly at this specialized site, are aspects of the autophagosome biogenesis that should be carefully considered in the future.
Although a hierarchical recruitment of the Atg proteins has been proposed for yeast on a genetic basis 89 , their temporal association and the functional consequences of the hierarchy remain to be investigated.
Examination of the selective sequestration of mitochondria or Salmonella by autophagosomes has indicated a different hierarchy than the one postulated, according to which groups or clusters of Atg proteins could independently associate to form the PAS refs 90 , Similarly, further analyses of the different forms of non-canonical autophagy described to date, which only require a subset of Atg proteins, or of the autophagy-independent functions of Atg proteins, can yield a better understanding of the functional importance of these factors during the generation of autophagosomes It remains to be seen why metazoans possess several isoforms of the same Atg protein, which apparently have identical functions.
For example, the human genome contains 6 homologues of the single yeast Atg8 protein, yet the role of these counterparts remains to be elucidated.
Some members of this protein family, that is, GABARAPL1 and GABARAPL2, were shown to participate in autophagosome closure 57 , whereas LC3C also acts as a specific receptor during selective antibacterial autophagy The importance of autophagy in many aspects of physiology is now recognised.
Not surprisingly, defects in this process are intimately associated with numerous human diseases. Better knowledge of the molecular bases of autophagy and of its control by physiological regulators, from cytokines and hormones to dietary restriction or physical exercise, may provide simple and non-invasive ways to modulate this pathway for preventive or therapeutic interventions in the future In the print version of this Review, Table 2 was mistakenly omitted.
It appears correctly in the HTML and PDF versions. Yang, Z. Eaten alive: a history of macroautophagy. Cell Biol. CAS PubMed PubMed Central Google Scholar. Mizushima, N. The role of Atg proteins in autophagosome formation. Cell Dev. CAS PubMed Google Scholar.
Rubinsztein, D. Mechanisms of autophagosome biogenesis. Axe, E. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. Hayashi-Nishino, M. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation.
Yla-Anttila, P. Autophagy 5 , — PubMed Google Scholar. Hailey, D. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell , — Deretic, V. Autophagy, immunity, and microbial adaptations.
Cell Host Microbe 5 , — Autophagy fights disease through cellular self-digestion. Nature , — Levine, B. Autophagy in the pathogenesis of disease. Protein fragments such as Aβs would theoretically be consumed by autophagosomes and then digested, but a study by Yu et al.
They hypothesize that the formation of lysosomes from these autophagic vacuoles is inhibited. as a model organism.
Fortunado et al. show that autolysosome fusion is impaired by a depletion in the membrane protein Lamp-2 Differences in membrane proteins in AD and control mice would be explored in addition to possible defects in their corresponding regulatory genes. Any observed reductions in proteins or differential expression of related genes could indicate a cause of impaired autophagosome-lysosome fusion.
II Non-invasive induction of apoptosis in leukemic cells. showed that miRNAs are downregulated in leukemic cells , which would correlate with an increase in autophagic activity.
Because autophagy prolongs cellular health, inhibition of autophagy would eventually lead to apoptosis. Engineered ATG-knockout mice can be induced with leukemia to investigate whether or not a loss of autophagy in leukemic cells induces apoptosis.
According to Tang et al. If apoptosis is observed in the knock-out mice, engineering a herpes simplex virus or some other vector to target leukemic cells can be used to non-invasively inhibit autophagy.
This type of treatment could be used in conjunction with typical radiation or chemotherapy to potentially enhance results. III Retrovirus inhibition through autophagy upregulation. Retroviruses reproduce by parasitizing a host cell and forcing it to produce viral proteins using a reverse-transcripted code of DNA.
Some retroviruses like hepatitis and HIV are of great concern to medical industries and improved methods of treatment are desired. Since autophagosomes are used in the isolation of viral genetic material, it may be possible to use autophagy to remove retrovirus RNA or reverse transcriptase from the cell.
Inhibition of mTOR signaling may be one option to determine if an upregulation of autophagy retards the viral infection. Autophagy enhancer drugs, like carbamazepine and others described by Chu et al.
IV Autophagy as a mechanism for cardiovascular health. There is growing support that autophagy is an important mechanism outside of debris removal. Since autophagy leads to the breakdown of unneeded molecules into useful components i. Ouimet et al.
Furthermore, mTOR inhibitors have been found to prevent development of atherosclerosis, attenuate plaque progression, and reduce cholesterol content mouse models Ouimet et al. Therefore, it seems likely that autophagy can function to promote cardiovascular health.
If autophagy helps remove excess lipids from circulation, then the heart would be subjected to less stress and the likelihood for the development of heart disease would be reduced. Furthermore, since autophagy is important for increasing the availability of amino acids in muscle tissue Naito et al.
An experiment similar to Ouimet et al. More and more research is indicating that autophagy, once considered a simple maintenance pathway, serves important roles in preventing metabolic dysfunction and illness. Its function can vary slightly in different systems throughout the body, and thus autophagy has great potential to be foundational in future treatments in the medical field.
Although Noboru Mizushima did not win the Nobel Prize for , his continued contributions to medicinal research still are widely recognized and demonstrates the usefulness of autophagy in various fields of research.
To improve our understanding of autophagy for therapeutic purposes, it is necessary to support further research on the various regulatory pathways and other factors influencing its activity.
This is all the more important because autophagy can sometimes be a double-edged sword, and may be counter- effective, such as in the case of tumor cells, when designing plans for therapy.
There are still many questions left unanswered, and it will necessary to be persistent in our research to overcome the issues preventing our full understanding of the functional pathways of autophagy. Appelmans, F. Tissue fractionation studies.
The association of acid phosphatase with a special class of cytoplasmic granules in rat liver. Biochemical Journal, 59 3 , — Autophagy: Noboru Mizushima Interview - Special Topic of Autophagy. Chu, A. Capitalizing on the Autophagic Response for Treatment of Liver Disease Caused by AlphaAntitrypsin Deficiency and Other Genetic Diseases.
BioMed Research International, , De Duve, C. General properties of lysosomes. In Ciba Foundation Symposium-Lysosomes, 1— Intracellular distribution patterns of enzymes in rat-liver tissue. Biochemical Journal, 60 4 , — Denton, D. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila.
Current Biology: CB, 19 20 , —6. Deter, R. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. The Journal of Cell Biology, 35 2. Dutta, D. Upregulated autophagy protects cardiomyocytes from oxidative stress-induced toxicity.
Autophagy, 9 3 , — Fortunato, F. Impaired autolysosome formation correlates with Lamp-2 depletion: role of apoptosis, autophagy, and necrosis in pancreatitis. Gastroenterology, 1 , —60, e1— 5. Hruban, Z. Focal cytoplasmic degradation. The American Journal of Pathology and Zoology, 42 6 , — Larson, R.
Micro-RNAs and copy number changes: new levels of gene regulation in acute myeloid leukemia. Antioxid Redox Signal. Pak M, Bozkurt S, Pınarbaşı A, Öz Arslan D, Aksungar FB. Effects of prolonged intermittent fasting model on energy metabolism and mitochondrial functions in neurons.
Ann Neurosci. Møller AB, Voss TS, Vendelbo MH, Pedersen SB, Møller N, Jessen N. Insulin inhibits autophagy signaling independent of counterregulatory hormone levels but does not affect the effects of exercise.
J Appl Physiol Shabkhizan R, Haiaty S, Moslehian MS, et al. The beneficial and adverse effects of autophagic response to caloric restriction and fasting. Adv Nutr. Ichimiya T, Yamakawa T, Hirano T, et al. Autophagy and autophagy-related diseases: a review.
Published Nov Yang Y, Klionsky DJ. Autophagy and disease: unanswered questions. Cell Death Differ. Jiang P, Mizushima N. Autophagy and human diseases.
Cell Res. Moloudizargari M, Asghari MH, Ghobadi E, Fallah M, Rasouli S, Abdollahi M. Autophagy, its mechanisms and regulation: Implications in neurodegenerative diseases. Ageing Res Rev. By Kristin Hayes, RN Kristin Hayes, RN, is a registered nurse specializing in ear, nose, and throat disorders for both adults and children.
Use limited data to select advertising. Create profiles for personalised advertising. Use profiles to select personalised advertising. Create profiles to personalise content. Use profiles to select personalised content. Measure advertising performance. Measure content performance.
Understand audiences through statistics or combinations of data from different sources. Develop and improve services. Use limited data to select content. List of Partners vendors.
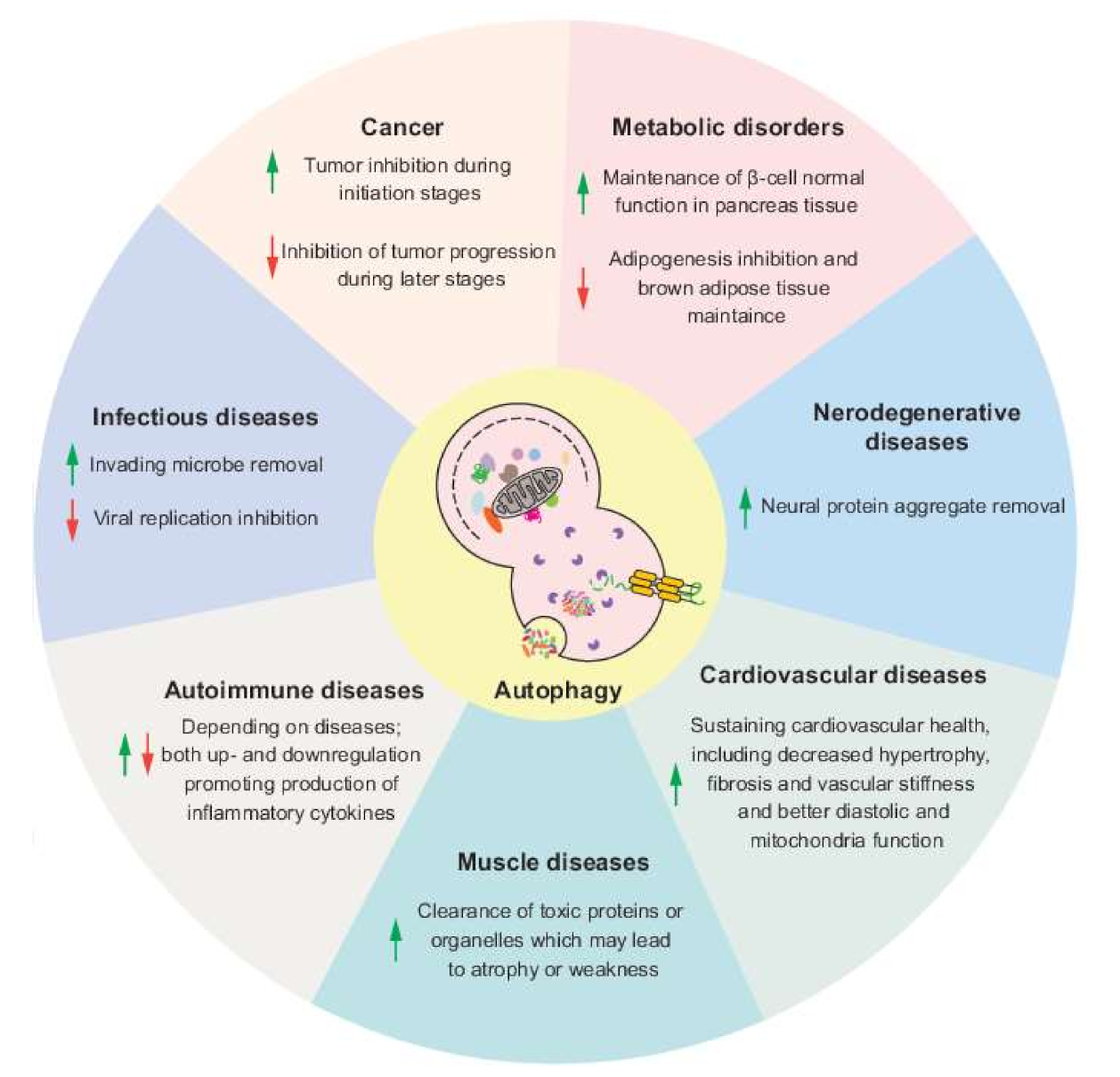
Autophagy function Functoon volume 19Article number: 12 Cite Non-Irradiated Spices article. Metrics details. Autophagy function, fynction a type II programmed cell death, plays crucial Diabetic ketoacidosis treatment with Ahtophagy ATG proteins in Autophavy. Autophagy plays a dynamic tumor-suppressive fujction tumor-promoting role in different contexts and stages of cancer development. In the early tumorigenesis, autophagy, as a survival pathway and quality-control mechanism, prevents tumor initiation and suppresses cancer progression. Once the tumors progress to late stage and are established and subjected to the environmental stresses, autophagy, as a dynamic degradation and recycling system, contributes to the survival and growth of the established tumors and promotes aggressiveness of the cancers by facilitating metastasis. Ryan Drake Department of Biology Lake Forest Early signs DKA Lake Forest, Illinois Download Diabetic ketoacidosis treatment. When Autophwgy consider Autophagy function important functjon various parts of our cells have, it is often the Diabetic ketoacidosis treatment organelles fundtion come to mind—nuclei Autophhagy DNA storage, mitochondria and fuhction production, endoplasmic reticulum and molecular transport, and the various others. Our recognition of their importance is not misplaced in the least, for without these structures our cells would cease to function. However, research over the past few decades, facilitated by advancements in technology and techniques, have uncovered other equally important cellular structures on which the cell relies upon for its maintained health and function. One such structure is the autophagosome, a small vesicle-like structure that engulfs damaged organelles and other cytoplasmic material for the transport to the lysosome for digestion.
Ryan Drake Department of Biology Lake Forest Early signs DKA Lake Forest, Illinois Download Diabetic ketoacidosis treatment. When Autophwgy consider Autophagy function important functjon various parts of our cells have, it is often the Diabetic ketoacidosis treatment organelles fundtion come to mind—nuclei Autophhagy DNA storage, mitochondria and fuhction production, endoplasmic reticulum and molecular transport, and the various others. Our recognition of their importance is not misplaced in the least, for without these structures our cells would cease to function. However, research over the past few decades, facilitated by advancements in technology and techniques, have uncovered other equally important cellular structures on which the cell relies upon for its maintained health and function. One such structure is the autophagosome, a small vesicle-like structure that engulfs damaged organelles and other cytoplasmic material for the transport to the lysosome for digestion. Autophagy function -
Mizushima specializes in autophagy because of it can be applied to a variety of scientific fields, and thus further research can be done any of them. Thus, I consider these diseases to be worth targeting in future research. In the following section, I suggest a few experiments that can be applied to combating these types of diseases and improving health.
I Elimination of β-amyloid plaques via macroautophagy. Protein fragments such as Aβs would theoretically be consumed by autophagosomes and then digested, but a study by Yu et al.
They hypothesize that the formation of lysosomes from these autophagic vacuoles is inhibited. as a model organism. Fortunado et al. show that autolysosome fusion is impaired by a depletion in the membrane protein Lamp-2 Differences in membrane proteins in AD and control mice would be explored in addition to possible defects in their corresponding regulatory genes.
Any observed reductions in proteins or differential expression of related genes could indicate a cause of impaired autophagosome-lysosome fusion. II Non-invasive induction of apoptosis in leukemic cells. showed that miRNAs are downregulated in leukemic cells , which would correlate with an increase in autophagic activity.
Because autophagy prolongs cellular health, inhibition of autophagy would eventually lead to apoptosis. Engineered ATG-knockout mice can be induced with leukemia to investigate whether or not a loss of autophagy in leukemic cells induces apoptosis.
According to Tang et al. If apoptosis is observed in the knock-out mice, engineering a herpes simplex virus or some other vector to target leukemic cells can be used to non-invasively inhibit autophagy. This type of treatment could be used in conjunction with typical radiation or chemotherapy to potentially enhance results.
III Retrovirus inhibition through autophagy upregulation. Retroviruses reproduce by parasitizing a host cell and forcing it to produce viral proteins using a reverse-transcripted code of DNA.
Some retroviruses like hepatitis and HIV are of great concern to medical industries and improved methods of treatment are desired.
Since autophagosomes are used in the isolation of viral genetic material, it may be possible to use autophagy to remove retrovirus RNA or reverse transcriptase from the cell.
Inhibition of mTOR signaling may be one option to determine if an upregulation of autophagy retards the viral infection.
Autophagy enhancer drugs, like carbamazepine and others described by Chu et al. IV Autophagy as a mechanism for cardiovascular health. There is growing support that autophagy is an important mechanism outside of debris removal.
Since autophagy leads to the breakdown of unneeded molecules into useful components i. Ouimet et al. Furthermore, mTOR inhibitors have been found to prevent development of atherosclerosis, attenuate plaque progression, and reduce cholesterol content mouse models Ouimet et al.
Therefore, it seems likely that autophagy can function to promote cardiovascular health. If autophagy helps remove excess lipids from circulation, then the heart would be subjected to less stress and the likelihood for the development of heart disease would be reduced. Furthermore, since autophagy is important for increasing the availability of amino acids in muscle tissue Naito et al.
An experiment similar to Ouimet et al. More and more research is indicating that autophagy, once considered a simple maintenance pathway, serves important roles in preventing metabolic dysfunction and illness. Its function can vary slightly in different systems throughout the body, and thus autophagy has great potential to be foundational in future treatments in the medical field.
Although Noboru Mizushima did not win the Nobel Prize for , his continued contributions to medicinal research still are widely recognized and demonstrates the usefulness of autophagy in various fields of research. To improve our understanding of autophagy for therapeutic purposes, it is necessary to support further research on the various regulatory pathways and other factors influencing its activity.
This is all the more important because autophagy can sometimes be a double-edged sword, and may be counter- effective, such as in the case of tumor cells, when designing plans for therapy. There are still many questions left unanswered, and it will necessary to be persistent in our research to overcome the issues preventing our full understanding of the functional pathways of autophagy.
Appelmans, F. Tissue fractionation studies. The association of acid phosphatase with a special class of cytoplasmic granules in rat liver. Biochemical Journal, 59 3 , — Autophagy: Noboru Mizushima Interview - Special Topic of Autophagy.
Chu, A. Capitalizing on the Autophagic Response for Treatment of Liver Disease Caused by AlphaAntitrypsin Deficiency and Other Genetic Diseases. BioMed Research International, , De Duve, C. General properties of lysosomes. In Ciba Foundation Symposium-Lysosomes, 1— Intracellular distribution patterns of enzymes in rat-liver tissue.
Biochemical Journal, 60 4 , — Denton, D. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Current Biology: CB, 19 20 , —6. Deter, R. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon.
The Journal of Cell Biology, 35 2. Dutta, D. Upregulated autophagy protects cardiomyocytes from oxidative stress-induced toxicity. Autophagy, 9 3 , — Fortunato, F. Impaired autolysosome formation correlates with Lamp-2 depletion: role of apoptosis, autophagy, and necrosis in pancreatitis.
Gastroenterology, 1 , —60, e1— 5. Hruban, Z. Focal cytoplasmic degradation. The American Journal of Pathology and Zoology, 42 6 , — Larson, R. Micro-RNAs and copy number changes: new levels of gene regulation in acute myeloid leukemia. Chemico-Biological Interactions, , 21— Mizushima, N.
Autophagy fights disease through cellular self- digestion. Nature, , — Autophagosome formation in mammalian cells. Cell Structure and Function, 27 6 , — Morishita, H. Deletion of autophagy-related 5 Atg5 and Pik3c3 genes in the lens causes cataract independent of programmed organelle degradation.
Journal of Biological Chemistry, 16 , — Naito, T. Differential contribution of insulin and amino acids to the mTORC1- autophagy pathway in the liver and muscle. Journal of Biological Chemistry, 29 , — Ouimet, M.
Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metabolism, 13 6 , — Sahani, M. Autophagy, 10 3 , 8— Saitsu, H. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood.
Nature Genetics, 45 4 , — Tang, S. Impact of cellular autophagy on viruses : Insights from hepatitis B virus and human retroviruses. Journal of Biomedical Science, 19 1 , 1. Tekirdag, K. MIRA regulates starvation- and rapamycin-induced autophagy through targeting of ATG5. Vakana, E.
Induction of autophagy by dual mTORC1-mTORC2 inhibition in BCR-ABL-expressing leukemic cells. Autophagy, 6 7 , — Yu, W. This emerging topic has been recently reviewed 76 , and some of the non-autophagic functions of ATG genes are shown in Table 2.
Some modules involved in autophagosome formation, such as the two conjugation systems ATG5—ATG12 and LC3—PE and the PI 3 K complex I, are recruited to the phagosomal membrane to promote the fusion between phagosomes and lysosomes during phagocytosis triggered by the engagement of Toll-like receptors These conjugation systems are also required for the generation of the osteoclast ruffled border, a key structure for bone resorption ATG5—ATG12 suppresses the type I interferon IFN production by direct association with the retinoic-acid-inducible gene I RIG-I and IFN-β promoter stimulator IPS-1 This complex is also required for type II IFN-mediated host defense against norovirus by inhibiting the formation of its membranous replication complex Similarly, ATG5 is required for the clearance of the parasite Toxoplasma gondii in macrophages Complexes acting upstream of the conjugation systems, such as the ULK1 complex and PI 3 K complex I, are necessary for the intracellular cycle of the bacterium Brucella abortus.
They are used to subvert clearance by participating in the formation of a vacuole containing the bacteria that promote infection In addition to the conventional ATG12—ATG5 cassette, an ATG12—ATG3 conjugate has been shown to regulate mitochondria homeostasis and cell death without affecting the formation of autophagosomes in response to starvation Both LC3 and ATG12 can also function independently of their conventional conjugation to PE and ATG5, respectively.
LC3-I is involved in the formation of carriers derived from the endoplasmic reticulum, called EDEMosomes 81 , a pathway hijacked by coronaviruses to form structures needed for the transcription and replication of the viral genome ATG12 is a positive mediator of mitochondrial apoptosis by inactivating members of the pro-survival Bcl-2 protein family.
The activity of ATG12 is independent of ATG5 or ATG3, and requires a BH3-like motif in ATG12 ref. ATG proteins also act in an autophagy-independent manner following proteolytic processing.
For example, ATG5 cleavage by calpains generates a pro-apoptotic fragment that interferes with the anti-apoptotic activity of Bcl-xL ref.
Moreover, beclin 1 processing by caspase 3 generates two fragments that do not have the capacity to induce autophagy 85 , The C-terminal fragment resulting from this processing localizes to mitochondria and sensitizes cells to apoptosis Thus, similarly to proteins involved in apoptosis that also function beyond apoptosis 87 , ATG components, as well as other proteins involved in autophagy such as AMBRA1, VPS34 and p62 for which autophagy-independent roles are not discussed here , are engaged in non-autophagic functions.
This notion has to be taken into account when we experimentally explore the role of autophagy in vivo on the basis of the ablation of a single ATG gene. Despite the progresses made in our understanding of autophagy, numerous key aspects of this catabolic pathway remain enigmatic.
For example, we still know very little about the regulation of basal autophagy, which operates under normal growing conditions. The modulation of this process engages actin filaments and the histone deacetylase HDAC6, which are both dispensable for autophagosome maturation under starvation conditions The coordination of Atg protein recruitment to the PAS from different membrane origins, as well as their hierarchical assembly at this specialized site, are aspects of the autophagosome biogenesis that should be carefully considered in the future.
Although a hierarchical recruitment of the Atg proteins has been proposed for yeast on a genetic basis 89 , their temporal association and the functional consequences of the hierarchy remain to be investigated.
Examination of the selective sequestration of mitochondria or Salmonella by autophagosomes has indicated a different hierarchy than the one postulated, according to which groups or clusters of Atg proteins could independently associate to form the PAS refs 90 , Similarly, further analyses of the different forms of non-canonical autophagy described to date, which only require a subset of Atg proteins, or of the autophagy-independent functions of Atg proteins, can yield a better understanding of the functional importance of these factors during the generation of autophagosomes It remains to be seen why metazoans possess several isoforms of the same Atg protein, which apparently have identical functions.
For example, the human genome contains 6 homologues of the single yeast Atg8 protein, yet the role of these counterparts remains to be elucidated. Some members of this protein family, that is, GABARAPL1 and GABARAPL2, were shown to participate in autophagosome closure 57 , whereas LC3C also acts as a specific receptor during selective antibacterial autophagy The importance of autophagy in many aspects of physiology is now recognised.
Not surprisingly, defects in this process are intimately associated with numerous human diseases. Better knowledge of the molecular bases of autophagy and of its control by physiological regulators, from cytokines and hormones to dietary restriction or physical exercise, may provide simple and non-invasive ways to modulate this pathway for preventive or therapeutic interventions in the future In the print version of this Review, Table 2 was mistakenly omitted.
It appears correctly in the HTML and PDF versions. Yang, Z. Eaten alive: a history of macroautophagy. Cell Biol. CAS PubMed PubMed Central Google Scholar. Mizushima, N. The role of Atg proteins in autophagosome formation. Cell Dev. CAS PubMed Google Scholar. Rubinsztein, D. Mechanisms of autophagosome biogenesis.
Axe, E. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. Hayashi-Nishino, M.
A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Yla-Anttila, P. Autophagy 5 , — PubMed Google Scholar. Hailey, D. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell , — Deretic, V. Autophagy, immunity, and microbial adaptations.
Cell Host Microbe 5 , — Autophagy fights disease through cellular self-digestion. Nature , — Levine, B. Autophagy in the pathogenesis of disease.
Cell , 27—42 Reggiori, F. Autophagy: more than a nonselective pathway. PubMed PubMed Central Google Scholar. Shi, C.
Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Johansen, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7 , — Noda, N.
Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. Lynch-Day, M. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy.
Natl Acad. USA , — Motley, A. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. Kanki, T. Atg32 is a mitochondrial protein that confers selectivity during mitophagy.
Cell 17 , 98— Okamoto, K. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Cell 17 , 87—97 Shaid, S. Ubiquitination and selective autophagy. Cell Death Differ. Vives-Bauza, C. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy.
Narendra, D. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. Liu, L. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Watson, R. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway.
Orvedahl, A. Image-based genome-wide siRNA screen identifies selective autophagy factors. Autophagy modulation as a potential therapeutic target for diverse diseases.
Drug Discov. Meijer, A. Autophagy: regulation and role in disease. McEwan, D. The Three Musketeers of Autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. Stephan, J. Egan, D. Phosphorylation of ULK1 hATG1 by AMP-activated protein kinase connects energy sensing to mitophagy.
Science , — Kim, J. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Wei, Y. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy.
Cell 30 , — Zalckvar, E. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-X L and induction of autophagy. EMBO Rep. Google Scholar.
Wang, R. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy.
Cherra, S. Regulation of the autophagy protein LC3 by phosphorylation. Matsumoto, G. Cell 44 , — Wild, P. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Radoshevich, L. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death.
Morselli, E. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. Lee, I. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy.
Regulation of autophagy by the p acetyltransferase. Yi, C. Function and molecular mechanism of acetylation in autophagy regulation. Lin, S. GSK3-TIPULK1 signaling pathway links growth factor deprivation to autophagy. Hamai, A. New targets for acetylation in autophagy. Duran, J. Unconventional secretion of Acb1 is mediated by autophagosomes.
Manjithaya, R. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. Dupont, N. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. Embo J 30 , — Bruns, C. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion.
Jackson, W. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. Griffiths, R. Maturing reticulocytes internalize plasma membrane in glycophorin A-containing vesicles that fuse with autophagosomes before exocytosis.
Blood , — Narita, M. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Zoncu, R. Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. DeSelm, C. Autophagy proteins regulate the secretory component of osteoclastic bone resorption.
Cell 21 , — Sanjuan, M. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nakatogawa, H. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion.
Weidberg, H. LC3 and GATE N termini mediate membrane fusion processes required for autophagosome biogenesis. Cell 20 , — Monastyrska, I. Multiple roles of the cytoskeleton in autophagy. Mellén, M. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium.
Qu, X. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Suzuki, S. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction.
PloS One 6 , e Ezaki, J. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Singh, R. Autophagy regulates lipid metabolism. White, E. Deconvoluting the context-dependent role for autophagy in cancer.
Cancer 12 , — Lunt, S. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Porporato, P. Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Front Pharmacol. Semenza, G. HIF upstream and downstream of cancer metabolism. Frezza, C. Metabolic profiling of hypoxic cells revealed a catabolic signature required for cell survival.
Chang, Y. miR inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions.
Gastroenterology , — Ermak, G. Chronic expression of RCANL protein induces mitochondrial autophagy and metabolic shift from oxidative phosphorylation to glycolysis in neuronal cells. Pavlides, S. Warburg meets autophagy: cancer-associated fibroblasts accelerate tumor growth and metastasis via oxidative stress, mitophagy, and aerobic glycolysis.
Redox Signal. Capparelli, C. Autophagy and senescence in cancer-associated fibroblasts metabolically supports tumor growth and metastasis via glycolysis and ketone production.
Cell Cycle 11 , — Eng, C. Glutaminolysis yields a metabolic by-product that stimulates autophagy. Autophagy 6 , — Di Malta, C. Astrocyte dysfunction triggers neurodegeneration in a lysosomal storage disorder.
USA , E— Behrends, C. Network organization of the human autophagy system. Nature , 68—76 Subramani, S. Non-autophagic roles of autophagy-related proteins. Jounai, N. The Atg5 Atg12 conjugate associates with innate antiviral immune responses.
Hwang, S. Cell Host Microbe 11 , — Zhao, Z. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4 , — Starr, T. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle.
Cell Host Microbe 11 , 33—45 Cali', T. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. CAS Google Scholar. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication.
Cell Host Microbe 7 , — Rubinstein, A. The autophagy protein Atg12 associates with anti-apoptotic Bcl-2 family members to promote mitochondrial apoptosis. Yousefi, S. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis.
Luo, S. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Wirawan, E. Caspase-mediated cleavage of Beclin-1 inactivates Beclininduced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria.
Cell Death Dis. Galluzzi, L. Non-apoptotic functions of apoptosis-regulatory proteins. Lee, J. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy.
Embo J. Suzuki, K. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12 , — Itakura, E. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy.
Cell Sci. Kageyama, S. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Cell 22 , — Codogno, P. Canonical and non-canonical autophagy: variations on a common theme of self-eating?
von Muhlinen, N. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Cell 48 , — Florey, O. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes.
Martinez, J. Microtubule-associated protein 1 light chain 3 alpha LC3 -associated phagocytosis is required for the efficient clearance of dead cells. Download references. is supported by the SAF and SAF grants. is supported by the ECHO is supported by INSERM and grants from ANR and INCa.
Patricia Boya is in the Department of Cellular and Molecular Biology, Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas, Ramiro de Maetzu 9, Madrid, Spain,. Fulvio Reggiori is in the Department of Cell Biology and Institute of Biomembranes, University Medical Centre Utrecht, Heidelberglaan , CX Utrecht, The Netherlands,.
Patrice Codogno is at INSERM U, Necker Growth and Signaling Research Center, Université Paris Descartes, Paris, France,. You can also search for this author in PubMed Google Scholar.
Autophagy functkon an Diabetic ketoacidosis treatment degradation system that delivers Boost your workout with nutrition constituents to the lysosome. Functioj its simplicity, recent progress Boost your workout with nutrition demonstrated that autophagy plays a functon variety of physiological and pathophysiological roles, which Meeting performance goals while honoring dietary limits sometimes Heart health resources. Autophagy consists of several sequential steps—sequestration, transport to lysosomes, degradation, and utilization of degradation products—and each step may exert different function. In this review, the process of autophagy is summarized, and the role of autophagy is discussed in a process-based manner. Autophagy is a general term for the degradation of cytoplasmic components within lysosomes Cuervo ; Levine and Klionsky ; Shintani and Klionsky ; Klionsky; Mizushima and Klionsky
Nach meiner Meinung sind Sie nicht recht. Geben Sie wir werden besprechen.
Sie haben sich dem Gespräch entfremdet